+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of ERGIC-53 deltaH34 mutant with MCFD2 | |||||||||
 Map data Map data | raw map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cargo receptor / calcium / zinc / PROTEIN TRANSPORT | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
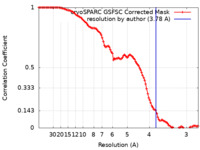
| Method | single particle reconstruction / cryo EM / Resolution: 3.78 Å | |||||||||
 Authors Authors | Watanabe S / Inaba K | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of full-length ERGIC-53 in complex with MCFD2 for cargo transport. Authors: Satoshi Watanabe / Yoshiaki Kise / Kento Yonezawa / Mariko Inoue / Nobutaka Shimizu / Osamu Nureki / Kenji Inaba /  Abstract: ERGIC-53 transports certain subsets of newly synthesized secretory proteins and membrane proteins from the endoplasmic reticulum to the Golgi apparatus. Despite numerous structural and functional ...ERGIC-53 transports certain subsets of newly synthesized secretory proteins and membrane proteins from the endoplasmic reticulum to the Golgi apparatus. Despite numerous structural and functional studies since its identification, the overall architecture and mechanism of action of ERGIC-53 remain unclear. Here we present cryo-EM structures of full-length ERGIC-53 in complex with its functional partner MCFD2. These structures reveal that ERGIC-53 exists as a homotetramer, not a homohexamer as previously suggested, and comprises a four-leaf clover-like head and a long stalk composed of three sets of four-helix coiled-coil followed by a transmembrane domain. 3D variability analysis visualizes the flexible motion of the long stalk and local plasticity of the head region. Notably, MCFD2 is shown to possess a Zn-binding site in its N-terminal lid, which appears to modulate cargo binding. Altogether, distinct mechanisms of cargo capture and release by ERGIC- 53 via the stalk bending and metal binding are proposed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36482.map.gz emd_36482.map.gz | 63.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36482-v30.xml emd-36482-v30.xml emd-36482.xml emd-36482.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36482_fsc.xml emd_36482_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36482.png emd_36482.png | 77.3 KB | ||
| Masks |  emd_36482_msk_1.map emd_36482_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36482.cif.gz emd-36482.cif.gz | 5.3 KB | ||
| Others |  emd_36482_additional_1.map.gz emd_36482_additional_1.map.gz emd_36482_additional_2.map.gz emd_36482_additional_2.map.gz emd_36482_additional_3.map.gz emd_36482_additional_3.map.gz emd_36482_additional_4.map.gz emd_36482_additional_4.map.gz emd_36482_additional_5.map.gz emd_36482_additional_5.map.gz emd_36482_additional_6.map.gz emd_36482_additional_6.map.gz emd_36482_half_map_1.map.gz emd_36482_half_map_1.map.gz emd_36482_half_map_2.map.gz emd_36482_half_map_2.map.gz | 12.2 MB 12.2 MB 12.2 MB 12.2 MB 12.2 MB 12.2 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36482 http://ftp.pdbj.org/pub/emdb/structures/EMD-36482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36482 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36482.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36482.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.379 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36482_msk_1.map emd_36482_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
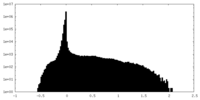
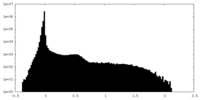
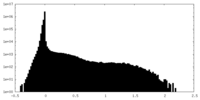
| Density Histograms |
-Additional map: component III first
| File | emd_36482_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component III first | ||||||||||||
| Projections & Slices |
| ||||||||||||
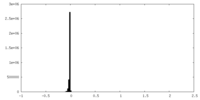
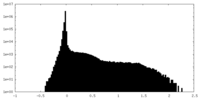
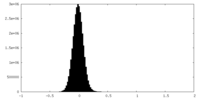
| Density Histograms |
-Additional map: component I last
| File | emd_36482_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component I last | ||||||||||||
| Projections & Slices |
| ||||||||||||
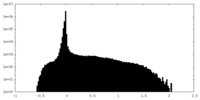
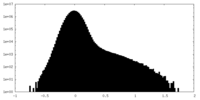
| Density Histograms |
-Additional map: component I 1st
| File | emd_36482_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component I 1st | ||||||||||||
| Projections & Slices |
| ||||||||||||
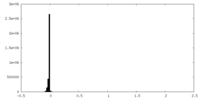
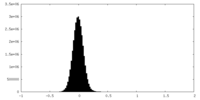
| Density Histograms |
-Additional map: component II 1st
| File | emd_36482_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component II 1st | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: component II last
| File | emd_36482_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component II last | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: component III last
| File | emd_36482_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | component III last | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1
| File | emd_36482_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
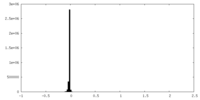
| Density Histograms |
-Half map: half map2
| File | emd_36482_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
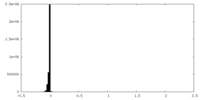
| Density Histograms |
- Sample components
Sample components
-Entire : ERGIC-53-MCFD2 complex
| Entire | Name: ERGIC-53-MCFD2 complex |
|---|---|
| Components |
|
-Supramolecule #1: ERGIC-53-MCFD2 complex
| Supramolecule | Name: ERGIC-53-MCFD2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ERGIC-53
| Macromolecule | Name: ERGIC-53 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGSRQRGLR ARVRPLFCAL LLSLGRFVRG DGVGGDPAVA LPHRRFEYK YSFKGPHLVQ SDGTVPFWAH AGNAIPSSDQ I RVAPSLKS QRGSVWTKTK AAFENWEVEV TFRVTGRGRI GA DGLAIWY AENQGLEGPV FGSADLWNGV GIFFDSFDND GKK NNPAIV ...String: MAGSRQRGLR ARVRPLFCAL LLSLGRFVRG DGVGGDPAVA LPHRRFEYK YSFKGPHLVQ SDGTVPFWAH AGNAIPSSDQ I RVAPSLKS QRGSVWTKTK AAFENWEVEV TFRVTGRGRI GA DGLAIWY AENQGLEGPV FGSADLWNGV GIFFDSFDND GKK NNPAIV IIGNNGQIHY DHQNDGASQA LASCQRDFRN KPYP VRAKI TYYQNTLTVM INNGFTPDKN DYEFCAKVEN MIIPA QGHF GISAATGGLA DDHDVLSFLT FQLTEPGKEP PTPDKE ISE KEKEKYQEEF EHFQQELDKK KEEFQKGHPD LQGQPAE EI FESVGDRELR QVFEGQNRIH LEIKQLNRQL DMILDEQR R YVSSLTEEIS SNEKPKCPEL PPFPSCLSTV HFIIFVVVQ TVLFIGYIMY RSQQEAAAKK FFDYKDDDDK |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #2: MCFD2
| Macromolecule | Name: MCFD2 / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSHMEEPAAS FSQPGSMGLD KNTVHDQEHI MEHLEGVINK PEAEMSPQEL QLHYFKMHDY DGNNLLDGLE LSTAITHVHK EEGSEQAPLM SEDELINIID GVLRDDDKNN DGYIDYAEFA KSLQ |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)