+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of isNS1 in complex with Fab56.2 and HDL | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flavivirus / Dengue / NS1 / HDL / VIRAL PROTEIN | |||||||||
| Biological species |  Dengue virus type 2 / Dengue virus type 2 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Chew BLA / Luo D | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Secreted dengue virus NS1 from infection is predominantly dimeric and in complex with high-density lipoprotein. Authors: Bing Liang Alvin Chew / A N Qi Ngoh / Wint Wint Phoo / Kitti Wing Ki Chan / Zheng Ser / Nikhil K Tulsian / Shiao See Lim / Mei Jie Grace Weng / Satoru Watanabe / Milly M Choy / Jenny Low / ...Authors: Bing Liang Alvin Chew / A N Qi Ngoh / Wint Wint Phoo / Kitti Wing Ki Chan / Zheng Ser / Nikhil K Tulsian / Shiao See Lim / Mei Jie Grace Weng / Satoru Watanabe / Milly M Choy / Jenny Low / Eng Eong Ooi / Christiane Ruedl / Radoslaw M Sobota / Subhash G Vasudevan / Dahai Luo /   Abstract: Severe dengue infections are characterized by endothelial dysfunction shown to be associated with the secreted nonstructural protein 1 (sNS1), making it an attractive vaccine antigen and ...Severe dengue infections are characterized by endothelial dysfunction shown to be associated with the secreted nonstructural protein 1 (sNS1), making it an attractive vaccine antigen and biotherapeutic target. To uncover the biologically relevant structure of sNS1, we obtained infection-derived sNS1 (isNS1) from dengue virus (DENV)-infected Vero cells through immunoaffinity purification instead of recombinant sNS1 (rsNS1) overexpressed in insect or mammalian cell lines. We found that isNS1 appeared as an approximately 250 kDa complex of NS1 and ApoA1 and further determined the cryoEM structures of isNS1 and its complex with a monoclonal antibody/Fab. Indeed, we found that the major species of isNS1 is a complex of the NS1 dimer partially embedded in a high-density lipoprotein (HDL) particle. Crosslinking mass spectrometry studies confirmed that the isNS1 interacts with the major HDL component ApoA1 through interactions that map to the NS1 wing and hydrophobic domains. Furthermore, our studies demonstrated that the sNS1 in sera from DENV-infected mice and a human patient form a similar complex as isNS1. Our results report the molecular architecture of a biological form of sNS1, which may have implications for the molecular pathogenesis of dengue. #2:  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Secreted dengue virus NS1 from infection is predominantly dimeric and in complex with high-density lipoprotein. Authors: Chew BLA / Ngoh AQ / Phoo WW / Chan KWK / Ser Z / Tulsian NK / Lim SS / Weng MJG / Watanabe S / Choy MM / Low JG / Ooi EE / Ruedl C / Sobota RM / Vasudevan SG / Luo D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36480.map.gz emd_36480.map.gz | 259.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36480-v30.xml emd-36480-v30.xml emd-36480.xml emd-36480.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
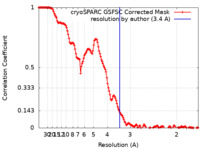
| FSC (resolution estimation) |  emd_36480_fsc.xml emd_36480_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36480.png emd_36480.png | 129.6 KB | ||
| Masks |  emd_36480_msk_1.map emd_36480_msk_1.map | 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36480.cif.gz emd-36480.cif.gz | 5.8 KB | ||
| Others |  emd_36480_half_map_1.map.gz emd_36480_half_map_1.map.gz emd_36480_half_map_2.map.gz emd_36480_half_map_2.map.gz | 255.1 MB 255.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36480 http://ftp.pdbj.org/pub/emdb/structures/EMD-36480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36480 | HTTPS FTP |
-Validation report
| Summary document |  emd_36480_validation.pdf.gz emd_36480_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36480_full_validation.pdf.gz emd_36480_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_36480_validation.xml.gz emd_36480_validation.xml.gz | 22.7 KB | Display | |
| Data in CIF |  emd_36480_validation.cif.gz emd_36480_validation.cif.gz | 29.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36480 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36480 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36480 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36480 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36480.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36480.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36480_msk_1.map emd_36480_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36480_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36480_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Fab56.2 with dengue virus nonstructural protei...
| Entire | Name: Ternary complex of Fab56.2 with dengue virus nonstructural protein 1 and High Density Lipoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Fab56.2 with dengue virus nonstructural protei...
| Supramolecule | Name: Ternary complex of Fab56.2 with dengue virus nonstructural protein 1 and High Density Lipoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: sNS1 derived from the supernatant of vero cells infected by DENV2 EDEN strain. Fab56.2 fragment was added in exogeneously, generated through proteolytic cleavage of antibody derived from hybridoma supernatant. |
|---|
-Supramolecule #2: isNS1ts
| Supramolecule | Name: isNS1ts / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Dengue virus 2 (EDEN2 strain, T164S mutant) secreted NS1 from infected vero cells |
|---|---|
| Source (natural) | Organism:  Dengue virus type 2 / Strain: EDEN2 Dengue virus type 2 / Strain: EDEN2 |
-Supramolecule #3: Monoclonal antibody fragment 56.2
| Supramolecule | Name: Monoclonal antibody fragment 56.2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: ab56.2 fragment was generated through proteolytic cleavage of antibody derived from hybridoma supernatant. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: core protein
| Macromolecule | Name: core protein / type: other / ID: 1 Details: secreted Non structural protein 1 from vero cell supernatant infected by dengue virus 2 Classification: other |
|---|---|
| Source (natural) | Organism:  Dengue virus type 2 / Strain: EDEN2 Dengue virus type 2 / Strain: EDEN2 |
| Sequence | String: DSGCVVSWKN KELKCGSGIF ITDNVHTWTE QYKFQPESPS KLASAIQKAH EEGICGIRSV TRLENLMWKQ ITPELNHILS ENEVKMTIMT GDIKGIMQAG KRSLRPQPTE LKYSWKAWGK AKMLSTELHN HTFLIDGPET AECPNTNRAW NSLEVEDYGF GVFTTNIWLK ...String: DSGCVVSWKN KELKCGSGIF ITDNVHTWTE QYKFQPESPS KLASAIQKAH EEGICGIRSV TRLENLMWKQ ITPELNHILS ENEVKMTIMT GDIKGIMQAG KRSLRPQPTE LKYSWKAWGK AKMLSTELHN HTFLIDGPET AECPNTNRAW NSLEVEDYGF GVFTTNIWLK LKERQDVSCD SKLMSAAIKD NRAVHADMGY WIESALNDTW KIEKASFIEV KSCHWPKSHT LWSNGVLESE MIIPKNFAGP VSQHNYRPGY HTQTAGPWHL GRLEMDFDFC EGTTVVVTED CGNRGPSLRT TTASGKLITE WCCRSCTLPP LRYRGEDGCW YGMEIRPLKE KEENLVNSLV TA GENBANK: GENBANK: EU081177 |
-Macromolecule #2: Fab 56.2 heavy chain
| Macromolecule | Name: Fab 56.2 heavy chain / type: other / ID: 2 / Details: heavy chain / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: ALEVQLQESG PGLVKPSQSL SFTWSVIGYS ITSGYYWKWI RQFSGNKLEW MGYIRYDGSN NYNPSLKNRI SIIRDTFKNQ FFLKLNFVTA GDTATYYCAN LYYYSSSPGW FPYWGQGTLV TVFAASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL ...String: ALEVQLQESG PGLVKPSQSL SFTWSVIGYS ITSGYYWKWI RQFSGNKLEW MGYIRYDGSN NYNPSLKNRI SIIRDTFKNQ FFLKLNFVTA GDTATYYCAN LYYYSSSPGW FPYWGQGTLV TVFAASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GPQTYICNVN HKPSNTKVDK KVEPKSCD |
-Macromolecule #3: Fab 56.2 light chain
| Macromolecule | Name: Fab 56.2 light chain / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: AELDIVMTQS PAIMSASLGE EITLTCSASS SVSYMHWYQQ KSGTSPKLLI YSTSNLASGV PSRFSGSGSG TFYSLTISSV EAEDAADYYC HQWSSWTFGG GTKLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD ...String: AELDIVMTQS PAIMSASLGE EITLTCSASS SVSYMHWYQQ KSGTSPKLLI YSTSNLASGV PSRFSGSGSG TFYSLTISSV EAEDAADYYC HQWSSWTFGG GTKLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT LSKADYEKHK VYACEVTHQG LSLPVTKSFN RGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X