+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ST1936-5HT6R complex | |||||||||
 Map data Map data | This is the cryo-EM density of ST1936-ht6 complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationadenylate cyclase-activating serotonin receptor signaling pathway / histamine receptor activity / Serotonin receptors / serotonin receptor activity / G protein-coupled serotonin receptor activity / neurotransmitter receptor activity / cerebral cortex cell migration / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation ...adenylate cyclase-activating serotonin receptor signaling pathway / histamine receptor activity / Serotonin receptors / serotonin receptor activity / G protein-coupled serotonin receptor activity / neurotransmitter receptor activity / cerebral cortex cell migration / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / positive regulation of TOR signaling / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / cellular response to glucagon stimulus / regulation of insulin secretion / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / platelet aggregation / G-protein beta/gamma-subunit complex binding / cognition / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / sensory perception of smell / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / cilium / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / dendrite / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Wen X / Sun J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2023 Journal: Biochem Biophys Res Commun / Year: 2023Title: Structural insight into the selective agonist ST1936 binding of serotonin receptor 5-HT6. Authors: Yuan Pei / Xin Wen / Sheng-Chao Guo / Zhi-Shuai Yang / Ru Zhang / Peng Xiao / Jin-Peng Sun /  Abstract: The serotonin receptor 5-HTR is an important G-protein-coupled receptor (GPCR) that involved in essential functions within the central and peripheral nervous systems and is linked to various ...The serotonin receptor 5-HTR is an important G-protein-coupled receptor (GPCR) that involved in essential functions within the central and peripheral nervous systems and is linked to various psychiatric disorders. Selective activation of 5-HTR promotes neural stem cell regeneration activity. As a 5-HTR selective agonist, 2-(5 chloro-2-methyl-1H-indol-3-yl)-N, N-dimethylethanolamine (ST1936) has been widely used to investigate the functions of the 5-HTR. The molecular mechanism of how ST1936 is recognized by 5-HTR and how it effectively couples with Gs remain unclear. Here, we reconstituted the ST1936-5-HTR-Gs complex in vitro and solved its cryo-electron microscopy structure at 3.1 Å resolution. Further structural analysis and mutational studies facilitated us to identify the residues of the Y310 and "toggle switch" W281 of the 5-HTR contributed to the higher efficacy of ST1936 compared with 5-HT. By uncovering the structural foundation of how 5-HTR specifically recognizes agonists and elucidating the molecular process of G protein activation, our discoveries offer valuable insights and pave the way for the development of promising 5-HTR agonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36409.map.gz emd_36409.map.gz | 27.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36409-v30.xml emd-36409-v30.xml emd-36409.xml emd-36409.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36409.png emd_36409.png | 20 KB | ||
| Filedesc metadata |  emd-36409.cif.gz emd-36409.cif.gz | 6.5 KB | ||
| Others |  emd_36409_half_map_1.map.gz emd_36409_half_map_1.map.gz emd_36409_half_map_2.map.gz emd_36409_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36409 http://ftp.pdbj.org/pub/emdb/structures/EMD-36409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36409 | HTTPS FTP |
-Validation report
| Summary document |  emd_36409_validation.pdf.gz emd_36409_validation.pdf.gz | 687.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36409_full_validation.pdf.gz emd_36409_full_validation.pdf.gz | 687.4 KB | Display | |
| Data in XML |  emd_36409_validation.xml.gz emd_36409_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_36409_validation.cif.gz emd_36409_validation.cif.gz | 12 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36409 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36409 | HTTPS FTP |
-Related structure data
| Related structure data |  8jlzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36409.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36409.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the cryo-EM density of ST1936-ht6 complex. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: This is the half1 map.
| File | emd_36409_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the half1 map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
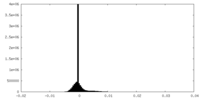
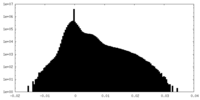
| Density Histograms |
-Half map: This is the half2 map.
| File | emd_36409_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the half2 map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
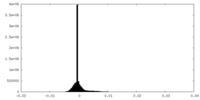
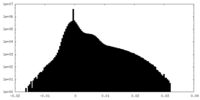
| Density Histograms |
- Sample components
Sample components
-Entire : st1936-HT6
| Entire | Name: st1936-HT6 |
|---|---|
| Components |
|
-Supramolecule #1: st1936-HT6
| Supramolecule | Name: st1936-HT6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Macromolecule #5: 5-hydroxytryptamine receptor 6
| Macromolecule | Name: 5-hydroxytryptamine receptor 6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.99209 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MVPEPGPTAN STPAWGAGPP SAPGGSGWVA AALCVVIALT AAANSLLIAL ICTQPALRNT SNFFLVSLFT SDLMVGLVVM PPAMLNALY GRWVLARGLC LLWTAFDVMC CSASILNLCL ISLDRYLLIL SPLRYKLRMT PLRALALVLG AWSLAALASF L PLLLGWHE ...String: MVPEPGPTAN STPAWGAGPP SAPGGSGWVA AALCVVIALT AAANSLLIAL ICTQPALRNT SNFFLVSLFT SDLMVGLVVM PPAMLNALY GRWVLARGLC LLWTAFDVMC CSASILNLCL ISLDRYLLIL SPLRYKLRMT PLRALALVLG AWSLAALASF L PLLLGWHE LGHARPPVPG QCRLLASLPF VLVASGLTFF LPSGAICFTY CRILLAARKQ AVQVASLTTG MASQASETLQ VP RTPRPGV ESADSRRLAT KHSRKALKAS LTLGILLGMF FVTWLPFFVA NIVQAVCDCI SPGLFDVLTW LGYCNSTMNP IIY PLFMRD FKRALGRFLP CPRCPRERQA SLASPSLRTS HSGPRPGLSL QQVLPLPLPP DSDSDSDAGS GGSSGLRLTA QLLL PGEAT QDPPLPTRAA AAVNFFNIDP AEPELRPHPL GIPTN UniProtKB: 5-hydroxytryptamine receptor 6 |
-Macromolecule #6: 2-(5-chloranyl-2-methyl-1~{H}-indol-3-yl)-N,N-dimethyl-ethanamine
| Macromolecule | Name: 2-(5-chloranyl-2-methyl-1~{H}-indol-3-yl)-N,N-dimethyl-ethanamine type: ligand / ID: 6 / Number of copies: 1 / Formula: UQL |
|---|---|
| Molecular weight | Theoretical: 236.74 Da |
| Chemical component information | 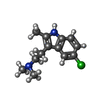 ChemComp-UQL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.09 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 286776 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)