+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CCHFV envelope protein Gc in complex with Gc13 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CCHFV / glycoprotein C / antibody / Bunyavirus / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / clathrin-dependent endocytosis of virus by host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Crimean-Congo hemorrhagic fever virus strain IbAr10200 / Crimean-Congo hemorrhagic fever virus strain IbAr10200 /  Crimean-Congo hemorrhagic fever orthonairovirus / Crimean-Congo hemorrhagic fever orthonairovirus /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Chong T / Cao S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus. Authors: Liushuai Li / Tingting Chong / Lu Peng / Yajie Liu / Guibo Rao / Yan Fu / Yanni Shu / Jiamei Shen / Qinghong Xiao / Jia Liu / Jiang Li / Fei Deng / Bing Yan / Zhihong Hu / Sheng Cao / Manli Wang /  Abstract: Crimean-Congo hemorrhagic fever virus (CCHFV) is a highly pathogenic tick-borne virus, prevalent in more than 30 countries worldwide. Human infection by this virus leads to severe illness, with an ...Crimean-Congo hemorrhagic fever virus (CCHFV) is a highly pathogenic tick-borne virus, prevalent in more than 30 countries worldwide. Human infection by this virus leads to severe illness, with an average case fatality of 40%. There is currently no approved vaccine or drug to treat the disease. Neutralizing antibodies are a promising approach to treat virus infectious diseases. This study generated 37 mouse-derived specific monoclonal antibodies against CCHFV Gc subunit. Neutralization assays using pseudotyped virus and authentic CCHFV identified Gc8, Gc13, and Gc35 as neutralizing antibodies. Among them, Gc13 had the highest neutralizing activity and binding affinity with CCHFV Gc. Consistently, Gc13, but not Gc8 or Gc35, showed in vivo protective efficacy (62.5% survival rate) against CCHFV infection in a lethal mouse infection model. Further characterization studies suggested that Gc8 and Gc13 may recognize a similar, linear epitope in domain II of CCHFV Gc, while Gc35 may recognize a different epitope in Gc. Cryo-electron microscopy of Gc-Fab complexes indicated that both Gc8 and Gc13 bind to the conserved fusion loop region and Gc13 had stronger interactions with sGc-trimers. This was supported by the ability of Gc13 to block CCHFV GP-mediated membrane fusion. Overall, this study provides new therapeutic strategies to treat CCHF and new insights into the interaction between antibodies with CCHFV Gc proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36407.map.gz emd_36407.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36407-v30.xml emd-36407-v30.xml emd-36407.xml emd-36407.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36407.png emd_36407.png | 70.6 KB | ||
| Filedesc metadata |  emd-36407.cif.gz emd-36407.cif.gz | 6 KB | ||
| Others |  emd_36407_half_map_1.map.gz emd_36407_half_map_1.map.gz emd_36407_half_map_2.map.gz emd_36407_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36407 http://ftp.pdbj.org/pub/emdb/structures/EMD-36407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36407 | HTTPS FTP |
-Validation report
| Summary document |  emd_36407_validation.pdf.gz emd_36407_validation.pdf.gz | 791.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36407_full_validation.pdf.gz emd_36407_full_validation.pdf.gz | 791.2 KB | Display | |
| Data in XML |  emd_36407_validation.xml.gz emd_36407_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  emd_36407_validation.cif.gz emd_36407_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36407 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36407 | HTTPS FTP |
-Related structure data
| Related structure data |  8jlxMC  8jkdC  8jlwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36407.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36407.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
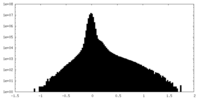
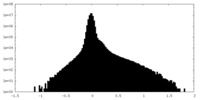
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36407_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
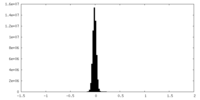
| Density Histograms |
-Half map: #1
| File | emd_36407_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
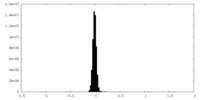
| Density Histograms |
- Sample components
Sample components
-Entire : CCHFV envelope protein Gc-Gc13 complex
| Entire | Name: CCHFV envelope protein Gc-Gc13 complex |
|---|---|
| Components |
|
-Supramolecule #1: CCHFV envelope protein Gc-Gc13 complex
| Supramolecule | Name: CCHFV envelope protein Gc-Gc13 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Crimean-Congo hemorrhagic fever virus strain IbAr10200 Crimean-Congo hemorrhagic fever virus strain IbAr10200 |
-Macromolecule #1: Glycoprotein C,CCHFV Gc fusion loops
| Macromolecule | Name: Glycoprotein C,CCHFV Gc fusion loops / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Crimean-Congo hemorrhagic fever orthonairovirus Crimean-Congo hemorrhagic fever orthonairovirus |
| Molecular weight | Theoretical: 65.880789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKNLLNSTSL ETSLSIEAPW GAINVQSTYK PTVSTANIAL SWSSVEHRGN KILVSGRSES IMKLEERTGI SWDLGVEDAS ESKLLTVSV MDLSQMYSPV FEYLSGDRQV GEWPKATCTG DCPERCGCTS STCLHKEWPH SRNWRCNPTW CWGVGTGCTC C GLDVKDLF ...String: MKNLLNSTSL ETSLSIEAPW GAINVQSTYK PTVSTANIAL SWSSVEHRGN KILVSGRSES IMKLEERTGI SWDLGVEDAS ESKLLTVSV MDLSQMYSPV FEYLSGDRQV GEWPKATCTG DCPERCGCTS STCLHKEWPH SRNWRCNPTW CWGVGTGCTC C GLDVKDLF TDYMFVKWKV EYIKTEAIVC VELTSQERQC SLIEAGTRFN LGPVTITLSE PRNIQQKLPP EIITLHPRIE EG FFDLMHV QKVLSASTVC KLQSCTHGVP GDLQVYHIGN LLKGDKVNGH LIHKIEPHFN TSWMSWDGCD LDYYCNMGDW PSC TYTGVT QHNHASFVNL LNIETDYTKN FHFHSKRVTA HGDTPQLDLK ARPTYGAGEI TVLVEVADME LHTKKIEISG LKFA SLACT GCYACSSGIS CKVRIHVDEP DELTVHVKSD DPDVVAASSS LMARKLEFGT DSTFKAFSAM PKTSLCFYIV EREHC KSCS EEDTKKCVNT KLEQPQSILI EHKGTIIGKQ NSTCTAKASR GSGGMKQIED KIEEILSKIY HIENEIARIK KLIGEG SGG SRGPFEGKPI PNPLLGLDST RTGHHHHHH UniProtKB: Envelopment polyprotein |
-Macromolecule #2: Mouse antibody Gc13 heavy chain
| Macromolecule | Name: Mouse antibody Gc13 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.931342 KDa |
| Sequence | String: QVQLQQSGPE VVRPGVSVKI SCKGSGYTFT DYATHWVKQS HVKSLEWIGV ISTYNGETDC NQKFKGKATM TVDKSSSTAY MELARLTSE DSAIYYCANG YYQAMDYWGQ GTSVTVSS |
-Macromolecule #3: Mouse antibody Gc13 light chain
| Macromolecule | Name: Mouse antibody Gc13 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.818041 KDa |
| Sequence | String: DIQMTQTPSS LSASLGDRVT ISCRASQDIS NYLNWYQQRP DGTLKLLIYY TSRLHSGVPS RFSGSGSGTD YSLTISNLEQ EDIATYFCQ QGSTLPWTFG GGTKLEIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 387809 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)