[English] 日本語
 Yorodumi
Yorodumi- EMDB-36285: Cryo-EM structure of human S1P transporter SPNS2 bound with an in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human S1P transporter SPNS2 bound with an inhibitor 16d | |||||||||
 Map data Map data | SPNS2 wild type full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transpoter / LIPID TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis ...regulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis / B cell homeostasis / transmembrane transporter activity / lymph node development / sensory perception of sound / bone development / endosome membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Pang B / Yu LY / Ren RB | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport. Authors: Bin Pang / Leiye Yu / Tong Li / Haizhan Jiao / Xiaomei Wu / Jinxin Wang / Ruiping He / Yurou Zhang / Juan Wang / Hongli Hu / Wei Dai / Li Chen / Ruobing Ren /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36285.map.gz emd_36285.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36285-v30.xml emd-36285-v30.xml emd-36285.xml emd-36285.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36285.png emd_36285.png | 41.8 KB | ||
| Filedesc metadata |  emd-36285.cif.gz emd-36285.cif.gz | 5.6 KB | ||
| Others |  emd_36285_half_map_1.map.gz emd_36285_half_map_1.map.gz emd_36285_half_map_2.map.gz emd_36285_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36285 http://ftp.pdbj.org/pub/emdb/structures/EMD-36285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36285 | HTTPS FTP |
-Related structure data
| Related structure data |  8jhrMC  8jhqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36285.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36285.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SPNS2 wild type full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
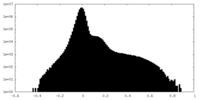
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: SPNS2 wild type half B map
| File | emd_36285_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SPNS2 wild type half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
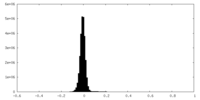
| Density Histograms |
-Half map: SPNS2 wild type half A map
| File | emd_36285_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SPNS2 wild type half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
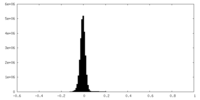
| Density Histograms |
- Sample components
Sample components
-Entire : Wild type SPNS2
| Entire | Name: Wild type SPNS2 |
|---|---|
| Components |
|
-Supramolecule #1: Wild type SPNS2
| Supramolecule | Name: Wild type SPNS2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sphingosine-1-phosphate transporter SPNS2
| Macromolecule | Name: Sphingosine-1-phosphate transporter SPNS2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.62193 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HSGDEVDAGS GHMMCLECAS AAAGGAEEEE ADAERRRRRR GAQRGAGGSG CCGARGAGGA GVSAAGDEVQ TLSGSVRRA PTGPPGTPGT PGCAATAKGP GAQQPKPASL GRGRGAAAAI LSLGNVLNYL DRYTVAGVLL DIQQHFGVKD R GAGLLQSV ...String: MHHHHHHHHH HSGDEVDAGS GHMMCLECAS AAAGGAEEEE ADAERRRRRR GAQRGAGGSG CCGARGAGGA GVSAAGDEVQ TLSGSVRRA PTGPPGTPGT PGCAATAKGP GAQQPKPASL GRGRGAAAAI LSLGNVLNYL DRYTVAGVLL DIQQHFGVKD R GAGLLQSV FICSFMVAAP IFGYLGDRFN RKVILSCGIF FWSAVTFSSS FIPQQYFWLL VLSRGLVGIG EASYSTIAPT II GDLFTKN TRTLMLSVFY FAIPLGSGLG YITGSSVKQA AGDWHWALRV SPVLGMITGT LILILVPATK RGHADQLGDQ LKA RTSWLR DMKALIRNRS YVFSSLATSA VSFATGALGM WIPLYLHRAQ VVQKTAETCN SPPCGAKDSL IFGAITCFTG FLGV VTGAG ATRWCRLKTQ RADPLVCAVG MLGSAIFICL IFVAAKSSIV GAYICIFVGE TLLFSNWAIT ADILMYVVIP TRRAT AVAL QSFTSHLLGD AGSPYLIGFI SDLIRQSTKD SPLWEFLSLG YALMLCPFVV VLGGMFFLAT ALFFVSDRAR AEQQVN QLA MPPASVKV UniProtKB: Sphingosine-1-phosphate transporter SPNS2 |
-Macromolecule #2: 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine
| Macromolecule | Name: 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine type: ligand / ID: 2 / Number of copies: 1 / Formula: YUX |
|---|---|
| Molecular weight | Theoretical: 343.506 Da |
| Chemical component information | 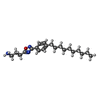 ChemComp-YUX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.52 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 113289 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)