[English] 日本語
 Yorodumi
Yorodumi- EMDB-36127: In situ structures of the ultra-long contracted tail of Myovirida... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structures of the ultra-long contracted tail of Myoviridae phage P1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Virus / Phage / VIRAL PROTEIN | |||||||||
| Function / homology | Gp22 Function and homology information Function and homology information | |||||||||
| Biological species |  Escherichia phage P1 (virus) Escherichia phage P1 (virus) | |||||||||
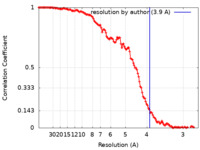
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhou JQ / Liu HR | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2023 Journal: Viruses / Year: 2023Title: In Situ Structures of the Ultra-Long Extended and Contracted Tail of Myoviridae Phage P1. Authors: Fan Yang / Liwen Wang / Junquan Zhou / Hao Xiao / Hongrong Liu /  Abstract: The phage tail is a common component of contractile injection systems (CISs), essential for exerting contractile function and facilitating membrane penetration of the inner tail tube. The near- ...The phage tail is a common component of contractile injection systems (CISs), essential for exerting contractile function and facilitating membrane penetration of the inner tail tube. The near-atomic resolution structures of the tail have been extensively studied, but the dynamic conformational changes before and after contraction and the associated molecular mechanism are still unclear. Here, we present the extended and contracted intact tail-structures of phage P1 by cryo-EM. The ultra-long tail of P1, 2450 Å in length, consists of a neck, a tail terminator, 53 repeated tail sheath rings, 53 repeated tube rings, and a baseplate. The sheath of the contracted tail shrinks by approximately 55%, resulting in the separation of the inner rigid tail tube from the sheath. The extended and contracted tails were further resolved by local reconstruction at 3.3 Å and 3.9 Å resolutions, respectively, allowing us to build the atomic models of the tail terminator protein gp24, the tube protein BplB, and the sheath protein gp22 for the extended tail, and of the sheath protein gp22 for the contracted tail. Our atomic models reveal the complex interaction network in the ultra-long tail and the novel conformational changes of the tail sheath between extended and contracted states. Our structures provide insights into the contraction and stabilization mechanisms of the tail. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36127.map.gz emd_36127.map.gz | 161.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36127-v30.xml emd-36127-v30.xml emd-36127.xml emd-36127.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36127_fsc.xml emd_36127_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_36127.png emd_36127.png | 132.6 KB | ||
| Filedesc metadata |  emd-36127.cif.gz emd-36127.cif.gz | 5.6 KB | ||
| Others |  emd_36127_half_map_1.map.gz emd_36127_half_map_1.map.gz emd_36127_half_map_2.map.gz emd_36127_half_map_2.map.gz | 138.7 MB 138.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36127 http://ftp.pdbj.org/pub/emdb/structures/EMD-36127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36127 | HTTPS FTP |
-Related structure data
| Related structure data |  8jajMC  8janC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36127.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36127.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
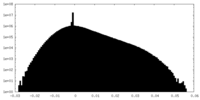
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
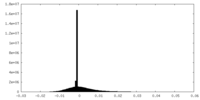
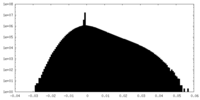
| Density Histograms |
-Half map: #1
| File | emd_36127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
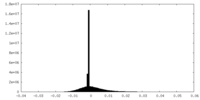
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage P1
| Entire | Name:  Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage P1
| Supramolecule | Name: Escherichia phage P1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2886926 / Sci species name: Escherichia phage P1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Gp22
| Macromolecule | Name: Gp22 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Molecular weight | Theoretical: 56.989246 KDa |
| Sequence | String: MSQYSIQQSL GNASGVAVSP INADATLSTG VALNSSLWAG IGVFARGKPF TVLAVTESNY EDVLGEPLKP SSGSQFEPIR HVYEAIQQT SGYVVRAVPD DAKFPIIMFD ESGEPAYSAL PYGSEIELDS GEAFAIYVDD GDPCISPTRE LTIETATADS A GNERFLLK ...String: MSQYSIQQSL GNASGVAVSP INADATLSTG VALNSSLWAG IGVFARGKPF TVLAVTESNY EDVLGEPLKP SSGSQFEPIR HVYEAIQQT SGYVVRAVPD DAKFPIIMFD ESGEPAYSAL PYGSEIELDS GEAFAIYVDD GDPCISPTRE LTIETATADS A GNERFLLK LTQTTSLGVV TTLETHTVSL AEEAKDDMGR LCYLPTALEA RSKYLRAVVN EELISTAKVT NKKSLAFTGG TN GDQSKIS TAAYLRAVKV LNNAPYMYTA VLGLGCYDNA AITALGKICA DRLIDGFFDV KPTLTYAEAL PAVEDTGLLG TDY VSCSVY HYPFSCKDKW TQSRVVFGLS GVAYAAKARG VKKNSDVGGW HYSPAGEERA VIARASIQPL YPEDTPDEEA MVKG RLNKV SVGTSGQMII DDALTCCTQD NYLHFQHVPS LMNAISRFFV QLARQMKHSP DGITAAGLTK GMTKLLDRFV ASGAL VAPR DPDADGTEPY VLKVTQAEFD KWEVVWACCP TGVARRIQGV PLLIK UniProtKB: Gp22 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)