+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SPARTA monomer bound with guide-target, state 2 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | SPARTA / Ago / Tir / DNA BINDING PROTEIN/DNA/RNA / DNA BINDING PROTEIN-DNA-RNA complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||
 Authors Authors | Li ZX / Guo LJ / Huang PP / Xiao YB / Chen MR | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system. Authors: Lijie Guo / Pingping Huang / Zhaoxing Li / Young-Cheul Shin / Purui Yan / Meiling Lu / Meirong Chen / Yibei Xiao /  Abstract: Short prokaryotic Ago accounts for most prokaryotic Argonaute proteins (pAgos) and is involved in defending bacteria against invading nucleic acids. Short pAgo associated with TIR-APAZ (SPARTA) has ...Short prokaryotic Ago accounts for most prokaryotic Argonaute proteins (pAgos) and is involved in defending bacteria against invading nucleic acids. Short pAgo associated with TIR-APAZ (SPARTA) has been shown to oligomerize and deplete NAD upon guide-mediated target DNA recognition. However, the molecular basis of SPARTA inhibition and activation remains unknown. In this study, we determined the cryogenic electron microscopy structures of Crenotalea thermophila SPARTA in its inhibited, transient and activated states. The SPARTA monomer is auto-inhibited by its acidic tail, which occupies the guide-target binding channel. Guide-mediated target binding expels this acidic tail and triggers substantial conformational changes to expose the Ago-Ago dimerization interface. As a result, SPARTA assembles into an active tetramer, where the four TIR domains are rearranged and packed to form NADase active sites. Together with biochemical evidence, our results provide a panoramic vision explaining SPARTA auto-inhibition and activation and expand understanding of pAgo-mediated bacterial defense systems. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36070.map.gz emd_36070.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36070-v30.xml emd-36070-v30.xml emd-36070.xml emd-36070.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36070.png emd_36070.png | 33 KB | ||
| Filedesc metadata |  emd-36070.cif.gz emd-36070.cif.gz | 6.5 KB | ||
| Others |  emd_36070_half_map_1.map.gz emd_36070_half_map_1.map.gz emd_36070_half_map_2.map.gz emd_36070_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36070 http://ftp.pdbj.org/pub/emdb/structures/EMD-36070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36070 | HTTPS FTP |
-Related structure data
| Related structure data |  8j8hMC  8j84C  8j9gC  8j9pC  8jayC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36070.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36070.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36070_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
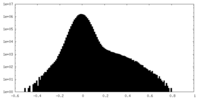
| Projections & Slices |
| ||||||||||||
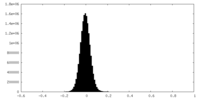
| Density Histograms |
-Half map: #1
| File | emd_36070_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
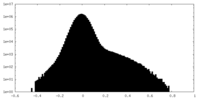
| Projections & Slices |
| ||||||||||||
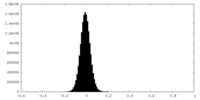
| Density Histograms |
- Sample components
Sample components
-Entire : Short ago complexed with TIR-APAZ
| Entire | Name: Short ago complexed with TIR-APAZ |
|---|---|
| Components |
|
-Supramolecule #1: Short ago complexed with TIR-APAZ
| Supramolecule | Name: Short ago complexed with TIR-APAZ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4, #1-#2 |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
-Macromolecule #1: Piwi domain-containing protein
| Macromolecule | Name: Piwi domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Molecular weight | Theoretical: 61.857793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMKELIY IEEPSILFAH GQKCTDPRDG LALFGPLNQI YGIKSGVVGT QKGLQIFKS YLDKIQKPIY NHNNITRPMF PGFEAVFGCK WESQNIVFKE ITDEEIRRYL FNASTHKRTY DLVTLFNDKI I TANKNDEE ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMKELIY IEEPSILFAH GQKCTDPRDG LALFGPLNQI YGIKSGVVGT QKGLQIFKS YLDKIQKPIY NHNNITRPMF PGFEAVFGCK WESQNIVFKE ITDEEIRRYL FNASTHKRTY DLVTLFNDKI I TANKNDEE RVDVWFVIVP EEIYKYCRPN SVLPNELVQT KSLISKSKAK SFRYTPTLFE EFNKKLKEVE KEAKTYNYDA QF HDQLKAR LLEHTIPTQI LRESTLAWRD FKNTFGAPIR DFSKIEGHLA WTISTAAYYK AGGKPWKLGD IRPGVCYLGL VYK KIEKSK NPQNACCAAQ MFLDNGDGTV FKGEVGPWYN PEKGEYHLKP KEAKALLTQA LESYKEQNKS YPKEVFIHAR TRFN DEEWN AFNEVTPKNT NLVGVTITKS KPLKLYKTEG AFPIMRGNAY IVDEKKAFLW TLGFVPKLQS TLSMEVPNPI FIEIN KGEA EIQQVLKDIL ALTKLNYNAC IYADGEPVTL RFANKIGEIL TASTEIKTPP LAFKYYI UniProtKB: Piwi domain-containing protein |
-Macromolecule #2: TIR domain-containing protein
| Macromolecule | Name: TIR domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Molecular weight | Theoretical: 56.809668 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMRNKIF ISHATPEDDD FTRWLSLKLI GLGYEVWCDI LFLDKGVDFW STIEKEIRE NTCKFLIVSS TAGNKREGVL KELAVATKVK KHLQDDMFII PLAIDENLSY DDINIEIVRL NAIDFKKSWA K GLQDLLDA ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMRNKIF ISHATPEDDD FTRWLSLKLI GLGYEVWCDI LFLDKGVDFW STIEKEIRE NTCKFLIVSS TAGNKREGVL KELAVATKVK KHLQDDMFII PLAIDENLSY DDINIEIVRL NAIDFKKSWA K GLQDLLDA FEKQNVPKKP PDHSKSNLLY QQIFLHDKQA IEKEETYDSN WFPIISFPNE LRFHRYDWRL PKQFDVRTLA FP AIRYKEY LCTFAWEYDF IHQLPKTETY NGQESIRIST SDILSGRYDT DFIRNYECQR LIVQLINKAF ELRMKDKNVR EYQ MSKTFA YWIEKGKLEK DKFEKIKLVG KQKNKYWHFG ISAAGKLYPS PVLMVSSHII FTMDGINLIK SKSIQHSSRR KQGK NWWND KWREKLLAFI RFLSDDQNAI YLNVGSEEKI LISNKPLKFF GKMSYVTPSE VTLEEESVLA DINNFEEDTE DLDEL EDIE UniProtKB: TIR domain-containing protein |
-Macromolecule #3: RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP...
| Macromolecule | Name: RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Molecular weight | Theoretical: 6.651949 KDa |
| Sequence | String: UGACGGCUCU AAUCUAUUAG U |
-Macromolecule #4: DNA (25-MER)
| Macromolecule | Name: DNA (25-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Molecular weight | Theoretical: 7.675 KDa |
| Sequence | String: (DC)(DA)(DA)(DC)(DT)(DA)(DA)(DT)(DA)(DG) (DA)(DT)(DT)(DA)(DG)(DA)(DG)(DC)(DC)(DG) (DT)(DC)(DA)(DA)(DT) |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 117819 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8j8h: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)