[English] 日本語
 Yorodumi
Yorodumi- EMDB-36051: Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determine... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | zinc / proton / transporter / Golgi apparatus / metal transporter / histidine-rich loop / METAL TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationzinc ion import into Golgi lumen / Golgi cis cisterna membrane / zinc ion transmembrane transporter activity / intracellular zinc ion homeostasis / sarcoplasmic reticulum membrane / cytoplasmic vesicle / vesicle / Golgi membrane / perinuclear region of cytoplasm / Golgi apparatus ...zinc ion import into Golgi lumen / Golgi cis cisterna membrane / zinc ion transmembrane transporter activity / intracellular zinc ion homeostasis / sarcoplasmic reticulum membrane / cytoplasmic vesicle / vesicle / Golgi membrane / perinuclear region of cytoplasm / Golgi apparatus / mitochondrion / identical protein binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | ||||||||||||
 Authors Authors | Han BB / Inaba K / Watanabe S | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn uptake into the Golgi apparatus. Authors: Han Ba Bui / Satoshi Watanabe / Norimichi Nomura / Kehong Liu / Tomoko Uemura / Michio Inoue / Akihisa Tsutsumi / Hiroyuki Fujita / Kengo Kinoshita / Yukinari Kato / So Iwata / Masahide Kikkawa / Kenji Inaba /  Abstract: Zinc ions (Zn) are vital to most cells, with the intracellular concentrations of Zn being tightly regulated by multiple zinc transporters located at the plasma and organelle membranes. We herein ...Zinc ions (Zn) are vital to most cells, with the intracellular concentrations of Zn being tightly regulated by multiple zinc transporters located at the plasma and organelle membranes. We herein present the 2.2-3.1 Å-resolution cryo-EM structures of a Golgi-localized human Zn/H antiporter ZnT7 (hZnT7) in Zn-bound and unbound forms. Cryo-EM analyses show that hZnT7 exists as a dimer via tight interactions in both the cytosolic and transmembrane (TM) domains of two protomers, each of which contains a single Zn-binding site in its TM domain. hZnT7 undergoes a TM-helix rearrangement to create a negatively charged cytosolic cavity for Zn entry in the inward-facing conformation and widens the luminal cavity for Zn release in the outward-facing conformation. An exceptionally long cytosolic histidine-rich loop characteristic of hZnT7 binds two Zn ions, seemingly facilitating Zn recruitment to the TM metal transport pathway. These structures permit mechanisms of hZnT7-mediated Zn uptake into the Golgi to be proposed. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36051.map.gz emd_36051.map.gz | 8.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36051-v30.xml emd-36051-v30.xml emd-36051.xml emd-36051.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36051_fsc.xml emd_36051_fsc.xml | 15.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_36051.png emd_36051.png | 46.3 KB | ||
| Masks |  emd_36051_msk_1.map emd_36051_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36051.cif.gz emd-36051.cif.gz | 6.7 KB | ||
| Others |  emd_36051_additional_1.map.gz emd_36051_additional_1.map.gz emd_36051_half_map_1.map.gz emd_36051_half_map_1.map.gz emd_36051_half_map_2.map.gz emd_36051_half_map_2.map.gz | 35.6 MB 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36051 http://ftp.pdbj.org/pub/emdb/structures/EMD-36051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36051 | HTTPS FTP |
-Related structure data
| Related structure data |  8j7wMC  8j7tC  8j7uC  8j7vC  8j7xC  8j7yC  8j80C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36051.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36051.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.788 Å | ||||||||||||||||||||||||||||||||||||
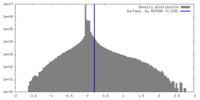
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36051_msk_1.map emd_36051_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
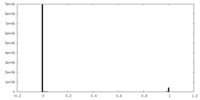
| Density Histograms |
-Additional map: #1
| File | emd_36051_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
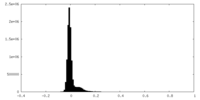
| Density Histograms |
-Half map: #2
| File | emd_36051_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
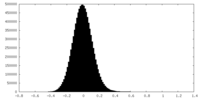
| Density Histograms |
-Half map: #1
| File | emd_36051_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human ZnT7-Fab complex
| Entire | Name: Human ZnT7-Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Human ZnT7-Fab complex
| Supramolecule | Name: Human ZnT7-Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Zinc transporter 7
| Macromolecule | Name: Zinc transporter 7 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.00432 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGGVAMPGAE DDVVMLPLSI KDDEYKPPKF NLFGKISGWF RSILSDKTSR NLFFFLCLNL SFAFVELLYG IWSNCLGLIS DSFHMFFDS TAILAGLAAS VISKWRDNDA FSYGYVRAEV LAGFVNGLFL IFTAFFIFSE GVERALAPPD VHHERLLLVS I LGFVVNLI ...String: MGGVAMPGAE DDVVMLPLSI KDDEYKPPKF NLFGKISGWF RSILSDKTSR NLFFFLCLNL SFAFVELLYG IWSNCLGLIS DSFHMFFDS TAILAGLAAS VISKWRDNDA FSYGYVRAEV LAGFVNGLFL IFTAFFIFSE GVERALAPPD VHHERLLLVS I LGFVVNLI GIFVFKHGGH GHSHGSGHGH SHSLFNGALD QAHGHVDHCH SHEVKHGAAH SHDHAHGHGH FHSHDGPSLK ET TGPSRQI LQGVFLHILA DTLGSIGVIA SAIMMQNFGL MIADPICSIL IAILIVVSVI PLLRESVGIL MQRTPPLLEN SLP QCYQRV QQLQGVYSLQ EQHFWTLCSD VYVGTLKLIV APDADARWIL SQTHNIFTQA GVRQLYVQID FAAM UniProtKB: Zinc transporter 7 |
-Macromolecule #2: Light chain of YN7114-08 Fab
| Macromolecule | Name: Light chain of YN7114-08 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.140529 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVLTQSPAS LAVSLRRRAT ISCRASESVD GYGHSFMHWY QQKSGQPPKL LIYRASNLES GVPARFSGSG SRTDFTLTID PVEADDAAT YYCQQSNEDP YTFGSGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLRRRAT ISCRASESVD GYGHSFMHWY QQKSGQPPKL LIYRASNLES GVPARFSGSG SRTDFTLTID PVEADDAAT YYCQQSNEDP YTFGSGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNRNEC |
-Macromolecule #3: Heavy chain of YN7114-08 Fab
| Macromolecule | Name: Heavy chain of YN7114-08 Fab / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.974102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQESGPG LVAPSQSLSI TCTVSGFSLT NYAVHWVRQS PGKGLEWLGV IWSNGRTDYN AAFISRLSIS KDNSKSQVFF KMNSLQADD TAIYYCARKL AYEGAMDYWG QGTSVTVSSA KTTPPSVYPL APGSAAQTNS MVTLGCLVKG YFPEPVTVTW N SGSLSSGV ...String: EVQLQESGPG LVAPSQSLSI TCTVSGFSLT NYAVHWVRQS PGKGLEWLGV IWSNGRTDYN AAFISRLSIS KDNSKSQVFF KMNSLQADD TAIYYCARKL AYEGAMDYWG QGTSVTVSSA KTTPPSVYPL APGSAAQTNS MVTLGCLVKG YFPEPVTVTW N SGSLSSGV HTFPAVLQSD LYTLSSSVTV PSSTWPSETV TCNVAHPASS TKVDKKIVPR DCGCKPCICT VPEVSS |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)