+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hydroxycarboxylic acid receptor / MK-6892 / Class A GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationneutrophil apoptotic process / nicotinic acid receptor activity / positive regulation of neutrophil apoptotic process / Hydroxycarboxylic acid-binding receptors / Class A/1 (Rhodopsin-like receptors) / positive regulation of adiponectin secretion / negative regulation of lipid catabolic process / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration ...neutrophil apoptotic process / nicotinic acid receptor activity / positive regulation of neutrophil apoptotic process / Hydroxycarboxylic acid-binding receptors / Class A/1 (Rhodopsin-like receptors) / positive regulation of adiponectin secretion / negative regulation of lipid catabolic process / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / GDP binding / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / cell junction / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Mao C / Gao M / Zang S / Zhu Y / Ma X / Zhang Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Orthosteric and allosteric modulation of human HCAR2 signaling complex. Authors: Chunyou Mao / Mengru Gao / Shao-Kun Zang / Yanqing Zhu / Dan-Dan Shen / Li-Nan Chen / Liu Yang / Zhiwei Wang / Huibing Zhang / Wei-Wei Wang / Qingya Shen / Yanhui Lu / Xin Ma / Yan Zhang /  Abstract: Hydroxycarboxylic acids are crucial metabolic intermediates involved in various physiological and pathological processes, some of which are recognized by specific hydroxycarboxylic acid receptors ...Hydroxycarboxylic acids are crucial metabolic intermediates involved in various physiological and pathological processes, some of which are recognized by specific hydroxycarboxylic acid receptors (HCARs). HCAR2 is one such receptor, activated by endogenous β-hydroxybutyrate (3-HB) and butyrate, and is the target for Niacin. Interest in HCAR2 has been driven by its potential as a therapeutic target in cardiovascular and neuroinflammatory diseases. However, the limited understanding of how ligands bind to this receptor has hindered the development of alternative drugs able to avoid the common flushing side-effects associated with Niacin therapy. Here, we present three high-resolution structures of HCAR2-Gi1 complexes bound to four different ligands, one potent synthetic agonist (MK-6892) bound alone, and the two structures bound to the allosteric agonist compound 9n in conjunction with either the endogenous ligand 3-HB or niacin. These structures coupled with our functional and computational analyses further our understanding of ligand recognition, allosteric modulation, and activation of HCAR2 and pave the way for the development of high-efficiency drugs with reduced side-effects. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36012.map.gz emd_36012.map.gz | 23.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36012-v30.xml emd-36012-v30.xml emd-36012.xml emd-36012.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36012.png emd_36012.png | 71.9 KB | ||
| Filedesc metadata |  emd-36012.cif.gz emd-36012.cif.gz | 7.1 KB | ||
| Others |  emd_36012_half_map_1.map.gz emd_36012_half_map_1.map.gz emd_36012_half_map_2.map.gz emd_36012_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36012 http://ftp.pdbj.org/pub/emdb/structures/EMD-36012 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36012 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36012 | HTTPS FTP |
-Related structure data
| Related structure data |  8j6rMC  8j6pC  8j6qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36012.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36012.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
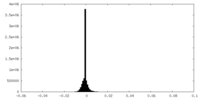
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36012_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
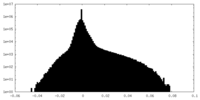
| Density Histograms |
-Half map: #2
| File | emd_36012_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
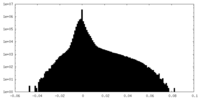
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex
| Entire | Name: Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex
| Supramolecule | Name: Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.225801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CTLSAEDKAA VERSKMIDRN LREDGEKAAR EVKLLLLGAG ESGKNTIVKQ MKIIHEAGYS EEECKQYKAV VYSNTIQSII AIIRAMGRL KIDFGDSARA DDARQLFVLA GAAEEGFMTA ELAGVIKRLW KDSGVQACFN RSREYQLNDS AAYYLNDLDR I AQPNYIPT ...String: CTLSAEDKAA VERSKMIDRN LREDGEKAAR EVKLLLLGAG ESGKNTIVKQ MKIIHEAGYS EEECKQYKAV VYSNTIQSII AIIRAMGRL KIDFGDSARA DDARQLFVLA GAAEEGFMTA ELAGVIKRLW KDSGVQACFN RSREYQLNDS AAYYLNDLDR I AQPNYIPT QQDVLRTRVK TTGIVETHFT FKDLHFKMFD VGAQRSERKK WIHCFEGVTA IIFCVALSDY DLVLAEDEEM NR MHASMKL FDSICNNKWF TDTSIILFLN KKDLFEEKIK KSPLTICYPE YAGSNTYEEA AAYIQCQFED LNKRKDTKEI YTH FTCSTD TKNVQFVFDA VTDVIIKNNL KDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: single Fab chain (scFv16)
| Macromolecule | Name: single Fab chain (scFv16) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.610615 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGS |
-Macromolecule #5: Hydroxycarboxylic acid receptor 2
| Macromolecule | Name: Hydroxycarboxylic acid receptor 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.366074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DHFLEIDKKN CCVFRDDFIV KVLPPVLGLE FIFGLLGNGL ALWIFCFHLK SWKSSRIFLF NLAVADFLLI ICLPFLMDNY VRRWDWKFG DIPCRLMLFM LAMNRQGSII FLTVVAVDRY FRVVHPHHAL NKISNRTAAI ISCLLWGITI GLTVHLLKKK M PIQNGGAN ...String: DHFLEIDKKN CCVFRDDFIV KVLPPVLGLE FIFGLLGNGL ALWIFCFHLK SWKSSRIFLF NLAVADFLLI ICLPFLMDNY VRRWDWKFG DIPCRLMLFM LAMNRQGSII FLTVVAVDRY FRVVHPHHAL NKISNRTAAI ISCLLWGITI GLTVHLLKKK M PIQNGGAN LCSSFSICHT FQWHEAMFLL EFFLPLGIIL FCSARIIWSL RQRQMDRHAK IKRAITFIMV VAIVFVICFL PS VVVRIRI FWLLHTSGTQ NCEVYRSVDL AFFITLSFTY MNSMLDPVVY YFSSPSFN UniProtKB: Hydroxycarboxylic acid receptor 2 |
-Macromolecule #6: 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-...
| Macromolecule | Name: 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: FI7 |
|---|---|
| Molecular weight | Theoretical: 386.402 Da |
| Chemical component information |  ChemComp-FI7: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)