+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of APOBEC3G-Vif complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human antiviral protein / HIV / ANTIVIRAL PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Kouno T / Shibata S / Hyun J / Kim TG / Wolf M | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G. Authors: Takahide Kouno / Satoshi Shibata / Megumi Shigematsu / Jaekyung Hyun / Tae Gyun Kim / Hiroshi Matsuo / Matthias Wolf /     Abstract: Great effort has been devoted to discovering the basis of A3G-Vif interaction, the key event of HIV's counteraction mechanism to evade antiviral innate immune response. Here we show reconstitution of ...Great effort has been devoted to discovering the basis of A3G-Vif interaction, the key event of HIV's counteraction mechanism to evade antiviral innate immune response. Here we show reconstitution of the A3G-Vif complex and subsequent A3G ubiquitination in vitro and report the cryo-EM structure of the A3G-Vif complex at 2.8 Å resolution using solubility-enhanced variants of A3G and Vif. We present an atomic model of the A3G-Vif interface, which assembles via known amino acid determinants. This assembly is not achieved by protein-protein interaction alone, but also involves RNA. The cryo-EM structure and in vitro ubiquitination assays identify an adenine/guanine base preference for the interaction and a unique Vif-ribose contact. This establishes the biological significance of an RNA ligand. Further assessment of interactions between A3G, Vif, and RNA ligands show that the A3G-Vif assembly and subsequent ubiquitination can be controlled by amino acid mutations at the interface or by polynucleotide modification, suggesting that a specific chemical moiety would be a promising pharmacophore to inhibit the A3G-Vif interaction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35998.map.gz emd_35998.map.gz | 48.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35998-v30.xml emd-35998-v30.xml emd-35998.xml emd-35998.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35998.png emd_35998.png | 111.1 KB | ||
| Others |  emd_35998_half_map_1.map.gz emd_35998_half_map_1.map.gz emd_35998_half_map_2.map.gz emd_35998_half_map_2.map.gz | 48.4 MB 48.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35998 http://ftp.pdbj.org/pub/emdb/structures/EMD-35998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35998 | HTTPS FTP |
-Validation report
| Summary document |  emd_35998_validation.pdf.gz emd_35998_validation.pdf.gz | 711.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35998_full_validation.pdf.gz emd_35998_full_validation.pdf.gz | 711.2 KB | Display | |
| Data in XML |  emd_35998_validation.xml.gz emd_35998_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_35998_validation.cif.gz emd_35998_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35998 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35998 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
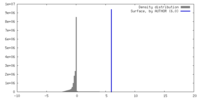
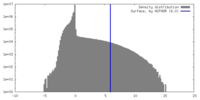
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35998_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
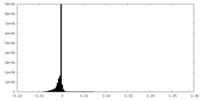
| Density Histograms |
-Half map: #2
| File | emd_35998_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
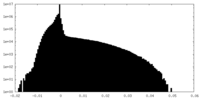
| Density Histograms |
- Sample components
Sample components
-Entire : APOBEC3G-Vif complex
| Entire | Name: APOBEC3G-Vif complex |
|---|---|
| Components |
|
-Supramolecule #1: APOBEC3G-Vif complex
| Supramolecule | Name: APOBEC3G-Vif complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sugar embedding | Material: Graphene oxide | ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: PLASMA CLEANING | ||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)