+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Agonist1 and Ruthenium Red bound state of mTRPV4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mTRPV4 / STRUCTURAL PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / negative regulation of brown fat cell differentiation / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production ...stretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / negative regulation of brown fat cell differentiation / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response / positive regulation of chemokine (C-X-C motif) ligand 1 production / positive regulation of chemokine (C-C motif) ligand 5 production / cartilage development involved in endochondral bone morphogenesis / cellular hypotonic salinity response / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / cellular hypotonic response / cortical microtubule organization / TRP channels / multicellular organismal-level water homeostasis / positive regulation of vascular permeability / osmosensory signaling pathway / cell-cell junction assembly / positive regulation of monocyte chemotactic protein-1 production / calcium ion import / cellular response to osmotic stress / response to osmotic stress / regulation of aerobic respiration / cortical actin cytoskeleton / diet induced thermogenesis / positive regulation of macrophage chemotaxis / microtubule polymerization / alpha-tubulin binding / beta-tubulin binding / monoatomic cation channel activity / energy homeostasis / cytoplasmic microtubule / SH2 domain binding / actin filament organization / protein kinase C binding / adherens junction / filopodium / positive regulation of JNK cascade / response to insulin / positive regulation of interleukin-6 production / calcium channel activity / ruffle membrane / intracellular calcium ion homeostasis / positive regulation of inflammatory response / actin filament binding / lamellipodium / glucose homeostasis / negative regulation of neuron projection development / cellular response to heat / growth cone / positive regulation of cytosolic calcium ion concentration / microtubule binding / response to hypoxia / calmodulin binding / positive regulation of ERK1 and ERK2 cascade / cilium / apical plasma membrane / focal adhesion / positive regulation of gene expression / lipid binding / protein kinase binding / cell surface / negative regulation of transcription by RNA polymerase II / ATP binding / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.88 Å | |||||||||
 Authors Authors | Zhen WX / Yang F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel. Authors: Wenxuan Zhen / Zhijun Zhao / Shenghai Chang / Xiaoying Chen / Yangzhuoqun Wan / Fan Yang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35922.map.gz emd_35922.map.gz | 51.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35922-v30.xml emd-35922-v30.xml emd-35922.xml emd-35922.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35922.png emd_35922.png | 45 KB | ||
| Filedesc metadata |  emd-35922.cif.gz emd-35922.cif.gz | 5.5 KB | ||
| Others |  emd_35922_half_map_1.map.gz emd_35922_half_map_1.map.gz emd_35922_half_map_2.map.gz emd_35922_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35922 http://ftp.pdbj.org/pub/emdb/structures/EMD-35922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35922 | HTTPS FTP |
-Validation report
| Summary document |  emd_35922_validation.pdf.gz emd_35922_validation.pdf.gz | 886.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35922_full_validation.pdf.gz emd_35922_full_validation.pdf.gz | 885.8 KB | Display | |
| Data in XML |  emd_35922_validation.xml.gz emd_35922_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_35922_validation.cif.gz emd_35922_validation.cif.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35922 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35922 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35922 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35922 | HTTPS FTP |
-Related structure data
| Related structure data |  8j1hMC  8j1bC  8j1dC  8j1fC  8jkmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35922.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35922.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35922_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35922_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mTRPV4 in complex with Agonist-1 and Ruthenium Red state
| Entire | Name: mTRPV4 in complex with Agonist-1 and Ruthenium Red state |
|---|---|
| Components |
|
-Supramolecule #1: mTRPV4 in complex with Agonist-1 and Ruthenium Red state
| Supramolecule | Name: mTRPV4 in complex with Agonist-1 and Ruthenium Red state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 4
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 4 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.666594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APAPQPPPIL KVFNRPILFD IVSRGSTADL DGLLSFLLTH KKRLTDEEFR EPSTGKTCLP KALLNLSNGR NDTIPVLLDI AERTGNMRE FINSPFRDIY YRGQTSLHIA IERRCKHYVE LLVAQGADVH AQARGRFFQP KDEGGYFYFG ELPLSLAACT N QPHIVNYL ...String: APAPQPPPIL KVFNRPILFD IVSRGSTADL DGLLSFLLTH KKRLTDEEFR EPSTGKTCLP KALLNLSNGR NDTIPVLLDI AERTGNMRE FINSPFRDIY YRGQTSLHIA IERRCKHYVE LLVAQGADVH AQARGRFFQP KDEGGYFYFG ELPLSLAACT N QPHIVNYL TENPHKKADM RRQDSRGNTV LHALVAIADN TRENTKFVTK MYDLLLLKCS RLFPDSNLET VLNNDGLSPL MM AAKTGKI GVFQHIIRRE VTDEDTRHLS RKFKDWAYGP VYSSLYDLSS LDTCGEEVSV LEILVYNSKI ENRHEMLAVE PIN ELLRDK WRKFGAVSFY INVVSYLCAM VIFTLTAYYQ PLEGTPPYPY RTTVDYLRLA GEVITLFTGV LFFFTSIKDL FTKK CPGVN SLFVDGSFQL LYFIYSVLVV VSAALYLAGI EAYLAVMVFA LVLGWMNALY FTRGLKLTGT YSIMIQKILF KDLFR FLLV YLLFMIGYAS ALVTLLNPCT NMKVCDEDQS NCTVPTYPAC RDSETFSAFL LDLFKLTIGM GDLEMLSSAK YPVVFI LLL VTYIILTFVL LLNMLIALMG ETVGQVSKES KHIWKLQWAT TILDIERSFP VFLRKAFRSG EMVTVGKSSD GTPDRRW CF RVDEVNWSHW NQNLGIINED PGK UniProtKB: Transient receptor potential cation channel subfamily V member 4 |
-Macromolecule #2: 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin...
| Macromolecule | Name: 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one type: ligand / ID: 2 / Number of copies: 4 / Formula: 9QM |
|---|---|
| Molecular weight | Theoretical: 448.465 Da |
| Chemical component information | 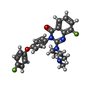 ChemComp-9QM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.88 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 428608 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)