[English] 日本語
 Yorodumi
Yorodumi- EMDB-35899: FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | FCP / heterodimer / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationlight-harvesting complex / photosynthesis, light harvesting in photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus / chloroplast / membrane Similarity search - Function | |||||||||
| Biological species |  Thalassiosira pseudonana (Diatom) Thalassiosira pseudonana (Diatom) | |||||||||
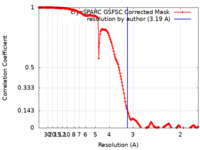
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Li Z / Feng Y / Wang W / Shen JR | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII. Authors: Yue Feng / Zhenhua Li / Xiaoyi Li / Lili Shen / Xueyang Liu / Cuicui Zhou / Jinyang Zhang / Min Sang / Guangye Han / Wenqiang Yang / Tingyun Kuang / Wenda Wang / Jian-Ren Shen /   Abstract: Diatoms rely on fucoxanthin chlorophyll -binding proteins (FCPs) for their great success in oceans, which have a great diversity in their pigment, protein compositions, and subunit organizations. We ...Diatoms rely on fucoxanthin chlorophyll -binding proteins (FCPs) for their great success in oceans, which have a great diversity in their pigment, protein compositions, and subunit organizations. We report a unique structure of photosystem II (PSII)-FCPII supercomplex from at 2.68-Å resolution by cryo-electron microscopy. FCPIIs within this PSII-FCPII supercomplex exist in dimers and monomers, and a homodimer and a heterodimer were found to bind to a PSII core. The FCPII homodimer is formed by Lhcf7 and associates with PSII through an Lhcx family antenna Lhcx6_1, whereas the heterodimer is formed by Lhcf6 and Lhcf11 and connects to the core together with an Lhcf5 monomer through Lhca2 monomer. An extended pigment network consisting of diatoxanthins, diadinoxanthins, fucoxanthins, and chlorophylls is revealed, which functions in efficient light harvesting, energy transfer, and dissipation. These results provide a structural basis for revealing the energy transfer and dissipation mechanisms and also for the structural diversity of FCP antennas in diatoms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35899.map.gz emd_35899.map.gz | 238.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35899-v30.xml emd-35899-v30.xml emd-35899.xml emd-35899.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35899_fsc.xml emd_35899_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35899.png emd_35899.png | 128.2 KB | ||
| Filedesc metadata |  emd-35899.cif.gz emd-35899.cif.gz | 6.3 KB | ||
| Others |  emd_35899_half_map_1.map.gz emd_35899_half_map_1.map.gz emd_35899_half_map_2.map.gz emd_35899_half_map_2.map.gz | 442.4 MB 442.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35899 http://ftp.pdbj.org/pub/emdb/structures/EMD-35899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35899 | HTTPS FTP |
-Related structure data
| Related structure data |  8j0dMC  8iwhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35899.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35899.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35899_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
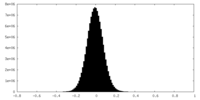
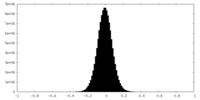
| Density Histograms |
-Half map: #1
| File | emd_35899_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
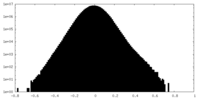
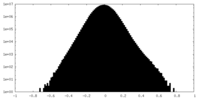
| Density Histograms |
- Sample components
Sample components
+Entire : M1 local refinement in the PSII-FCPII from the Thalassiosira pseu...
+Supramolecule #1: M1 local refinement in the PSII-FCPII from the Thalassiosira pseu...
+Macromolecule #1: Fucoxanthin chl a/c protein, lhca clade
+Macromolecule #2: Fucoxanthin-chlorophyll a-c binding protein, plastid
+Macromolecule #3: Fucoxanthin chlorophyll a/c protein 6
+Macromolecule #4: Fucoxanthin chlorophyll a/c protein 5
+Macromolecule #5: CHLOROPHYLL A
+Macromolecule #6: Chlorophyll c1
+Macromolecule #7: (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5...
+Macromolecule #8: (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,be...
+Macromolecule #9: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #10: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #11: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #12: Chlorophyll c2
+Macromolecule #13: DODECYL-BETA-D-MALTOSIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)