[English] 日本語
 Yorodumi
Yorodumi- EMDB-35890: Cellular components at INS-1E cell interior under first phase of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cellular components at INS-1E cell interior under first phase of glucose-stimulated insulin secretion | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | insulin secretion / cytoskeleton / CYTOSOLIC PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Li W / Li A / Yu B / Zhang X / Liu X / White K / Stevens R / Baumeister W / Sali A / Jasnin M / Sun L | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: In situ structure of actin remodeling during glucose-stimulated insulin secretion using cryo-electron tomography. Authors: Weimin Li / Angdi Li / Bing Yu / Xiaoxiao Zhang / Xiaoyan Liu / Kate L White / Raymond C Stevens / Wolfgang Baumeister / Andrej Sali / Marion Jasnin / Liping Sun /    Abstract: Actin mediates insulin secretion in pancreatic β-cells through remodeling. Hampered by limited resolution, previous studies have offered an ambiguous depiction as depolymerization and ...Actin mediates insulin secretion in pancreatic β-cells through remodeling. Hampered by limited resolution, previous studies have offered an ambiguous depiction as depolymerization and repolymerization. We report the in situ structure of actin remodeling in INS-1E β-cells during glucose-stimulated insulin secretion at nanoscale resolution. After remodeling, the actin filament network at the cell periphery exhibits three marked differences: 12% of actin filaments reorient quasi-orthogonally to the ventral membrane; the filament network mainly remains as cell-stabilizing bundles but partially reconfigures into a less compact arrangement; actin filaments anchored to the ventral membrane reorganize from a "netlike" to a "blooming" architecture. Furthermore, the density of actin filaments and microtubules around insulin secretory granules decreases, while actin filaments and microtubules become more densely packed. The actin filament network after remodeling potentially precedes the transport and release of insulin secretory granules. These findings advance our understanding of actin remodeling and its role in glucose-stimulated insulin secretion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35890.map.gz emd_35890.map.gz | 504.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35890-v30.xml emd-35890-v30.xml emd-35890.xml emd-35890.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35890.png emd_35890.png | 178 KB | ||
| Filedesc metadata |  emd-35890.cif.gz emd-35890.cif.gz | 3.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35890 http://ftp.pdbj.org/pub/emdb/structures/EMD-35890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35890 | HTTPS FTP |
-Validation report
| Summary document |  emd_35890_validation.pdf.gz emd_35890_validation.pdf.gz | 472.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35890_full_validation.pdf.gz emd_35890_full_validation.pdf.gz | 472.1 KB | Display | |
| Data in XML |  emd_35890_validation.xml.gz emd_35890_validation.xml.gz | 4.2 KB | Display | |
| Data in CIF |  emd_35890_validation.cif.gz emd_35890_validation.cif.gz | 4.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35890 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35890 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35890.map.gz / Format: CCP4 / Size: 544 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35890.map.gz / Format: CCP4 / Size: 544 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 12.112 Å | ||||||||||||||||||||||||||||||||
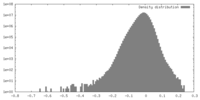
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : INS-1E
| Entire | Name: INS-1E |
|---|---|
| Components |
|
-Supramolecule #1: INS-1E
| Supramolecule | Name: INS-1E / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.45 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.5 / Focused ion beam - Duration: 30 / Focused ion beam - Temperature: 78 K / Focused ion beam - Initial thickness: 1000 / Focused ion beam - Final thickness: 180 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is FEI Aquilos2. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
- Image processing
Image processing
| Final reconstruction | Number images used: 61 |
|---|
 Movie
Movie Controller
Controller


















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)