+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Multidrug resistance-associated protein 3 | |||||||||
 Map data Map data | app mrp2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP-binding cassette transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucuronoside transmembrane transporter activity / icosanoid transmembrane transporter activity / ABC-type bile acid transporter activity / leukotriene transport / ABC-type glutathione S-conjugate transporter activity / ABC-type glutathione-S-conjugate transporter / : / xenobiotic transmembrane transport / Paracetamol ADME / NFE2L2 regulating MDR associated enzymes ...glucuronoside transmembrane transporter activity / icosanoid transmembrane transporter activity / ABC-type bile acid transporter activity / leukotriene transport / ABC-type glutathione S-conjugate transporter activity / ABC-type glutathione-S-conjugate transporter / : / xenobiotic transmembrane transport / Paracetamol ADME / NFE2L2 regulating MDR associated enzymes / ABC-type xenobiotic transporter / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / ABC-type xenobiotic transporter activity / xenobiotic transport / bile acid and bile salt transport / Aspirin ADME / xenobiotic transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transport across blood-brain barrier / Recycling of bile acids and salts / xenobiotic metabolic process / basal plasma membrane / ABC-family proteins mediated transport / transmembrane transport / basolateral plasma membrane / ciliary basal body / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
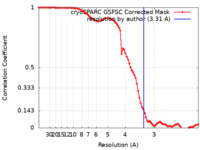
| Method | single particle reconstruction / cryo EM / Resolution: 3.31 Å | |||||||||
 Authors Authors | Yun CH / Gao HM | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: structure of multidrug resistance-associated protein 3 at 3.31 Angstroms Authors: Yun CH / Gao HM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35867.map.gz emd_35867.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35867-v30.xml emd-35867-v30.xml emd-35867.xml emd-35867.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35867_fsc.xml emd_35867_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_35867.png emd_35867.png | 97.8 KB | ||
| Filedesc metadata |  emd-35867.cif.gz emd-35867.cif.gz | 5.9 KB | ||
| Others |  emd_35867_half_map_1.map.gz emd_35867_half_map_1.map.gz emd_35867_half_map_2.map.gz emd_35867_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35867 http://ftp.pdbj.org/pub/emdb/structures/EMD-35867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35867 | HTTPS FTP |
-Validation report
| Summary document |  emd_35867_validation.pdf.gz emd_35867_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35867_full_validation.pdf.gz emd_35867_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35867_validation.xml.gz emd_35867_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_35867_validation.cif.gz emd_35867_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35867 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35867 | HTTPS FTP |
-Related structure data
| Related structure data |  8izpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35867.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35867.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | app mrp2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_35867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_35867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : multidrug resistance-associated protein 3 in apo state
| Entire | Name: multidrug resistance-associated protein 3 in apo state |
|---|---|
| Components |
|
-Supramolecule #1: multidrug resistance-associated protein 3 in apo state
| Supramolecule | Name: multidrug resistance-associated protein 3 in apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-binding cassette sub-family C member 3
| Macromolecule | Name: ATP-binding cassette sub-family C member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 174.547766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDALCGSGEL GSKFWDSNLS VHTENPDLTP CFQNSLLAWV PCIYLWVALP CYLLYLRHHC RGYIILSHLS KLKMVLGVLL WCVSWADLF YSFHGLVHGR APAPVFFVTP LVVGVTMLLA TLLIQYERLQ GVQSSGVLII FWFLCVVCAI VPFRSKILLA K AEGEISDP ...String: MDALCGSGEL GSKFWDSNLS VHTENPDLTP CFQNSLLAWV PCIYLWVALP CYLLYLRHHC RGYIILSHLS KLKMVLGVLL WCVSWADLF YSFHGLVHGR APAPVFFVTP LVVGVTMLLA TLLIQYERLQ GVQSSGVLII FWFLCVVCAI VPFRSKILLA K AEGEISDP FRFTTFYIHF ALVLSALILA CFREKPPFFS AKNVDPNPYP ETSAGFLSRL FFWWFTKMAI YGYRHPLEEK DL WSLKEED RSQMVVQQLL EAWRKQEKQT ARHKASAAPG KNASGEDEVL LGARPRPRKP SFLKALLATF GSSFLISACF KLI QDLLSF INPQLLSILI RFISNPMGPS WWGFLVAGLM FLCSMMQSLI LQHYYHYIFV TGVKFRTGIM GVIYRKALVI TNSV KRAST VGEIVNLMSV DAQRFMDLAP FLNLLWSAPL QIILAIYFLW QNLGPSVLAG VAFMVLLIPL NGAVAVKMRA FQVKQ MKLK DSRIKLMSEI LNGIKVLKLY AWEPSFLKQV EGIRQGELQL LRTAAYLHTT TTFTWMCSPF LVTLITLWVY VYVDPN NVL DAEKAFVSVS LFNILRLPLN MLPQLISNLT QASVSLKRIQ QFLSQEELDP QSVERKTISP GYAITIHSGT FTWAQDL PP TLHSLDIQVP KGALVAVVGP VGCGKSSLVS ALLGEMEKLE GKVHMKGSVA YVPQQAWIQN CTLQENVLFG KALNPKRY Q QTLEACALLA DLEMLPGGDQ TEIGEKGINL SGGQRQRVSL ARAVYSDADI FLLDDPLSAV DSHVAKHIFD HVIGPEGVL AGKTRVLVTH GISFLPQTDF IIVLADGQVS EMGPYPALLQ RNGSFANFLC NYAPDEDQGH LEDSWTALEG AEDKEALLIE DTLSNHTDL TDNDPVTYVV QKQFMRQLSA LSSDGEGQGR PVPRRHLGPS EKVQVTEAKA DGALTQEEKA AIGTVELSVF W DYAKAVGL CTTLAICLLY VGQSAAAIGA NVWLSAWTND AMADSRQNNT SLRLGVYAAL GILQGFLVML AAMAMAAGGI QA ARVLHQA LLHNKIRSPQ SFFDTTPSGR ILNCFSKDIY VVDEVLAPVI LMLLNSFFNA ISTLVVIMAS TPLFTVVILP LAV LYTLVQ RFYAATSRQL KRLESVSRSP IYSHFSETVT GASVIRAYNR SRDFEIISDT KVDANQRSCY PYIISNRWLS IGVE FVGNC VVLFAALFAV IGRSSLNPGL VGLSVSYSLQ VTFALNWMIR MMSDLESNIV AVERVKEYSK TETEAPWVVE GSRPP EGWP PRGEVEFRNY SVRYRPGLDL VLRDLSLHVH GGEKVGIVGR TGAGKSSMTL CLFRILEAAK GEIRIDGLNV ADIGLH DVR SQLTIIPQDP ILFSGTLRMN LDPFGSYSEE DIWWALELSH LHTFVSSQPA GLDFQCSEGG ENLSVGQRQL VCLARAL LR KSRILVLDEA TAAIDLETDN LIQATIRTQF DTCTVLTIAH RLNTIMDYTR VLVLDKGVVA EFDSPANLIA ARGIFYGM A RDAGLALEEN LYFQGSGGGG GGDYKDHDGD YKDHDIDYKD DDDKHHHHHH UniProtKB: ATP-binding cassette sub-family C member 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.00 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)