+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
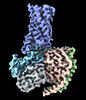
| Title | Lysophosphatidylserine receptor GPR174-Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbioactive lipid receptor activity / negative regulation of interleukin-2 production / T cell homeostasis / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis ...bioactive lipid receptor activity / negative regulation of interleukin-2 production / T cell homeostasis / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / G protein-coupled receptor activity / bone development / platelet aggregation / centriolar satellite / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / intracellular membrane-bounded organelle / GTPase activity / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Gong W / Liu G / Li X / Zhang X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174. Authors: Guibing Liu / Xiu Li / Yujing Wang / Xuan Zhang / Weimin Gong /   Abstract: Lysophosphatidylserine (LysoPS) is a naturally occurring lipid mediator involved in various physiological and pathological processes especially those related to the immune system. GPR34, GPR174, and ...Lysophosphatidylserine (LysoPS) is a naturally occurring lipid mediator involved in various physiological and pathological processes especially those related to the immune system. GPR34, GPR174, and P2Y10 have been identified as the receptors for LysoPS, and its analogues have been developed as agonists or antagonists for these receptors. However, the lack of structural information hinders the drug development with novel characteristics, such as nonlipid ligands and allosteric modulators. Here, we determined the structures of human GPR34 and GPR174 in complex with LysoPS and G protein by cryo-EM. Combined with structural analysis and functional studies, we elucidated the lipid-binding modes of these receptors. By structural comparison, we identified the structural features of GPR34 and GPR174 in active state. Taken together, our findings provide insights into ligand recognition and signaling of LysoPS receptors and will facilitate the development of novel therapeutics for related inflammatory diseases and autoimmune diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35838.map.gz emd_35838.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35838-v30.xml emd-35838-v30.xml emd-35838.xml emd-35838.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35838.png emd_35838.png | 126.7 KB | ||
| Filedesc metadata |  emd-35838.cif.gz emd-35838.cif.gz | 6.5 KB | ||
| Others |  emd_35838_half_map_1.map.gz emd_35838_half_map_1.map.gz emd_35838_half_map_2.map.gz emd_35838_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35838 http://ftp.pdbj.org/pub/emdb/structures/EMD-35838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35838 | HTTPS FTP |
-Related structure data
| Related structure data |  8izbMC  8wrbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35838.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35838.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35838_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
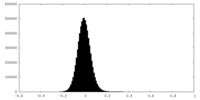
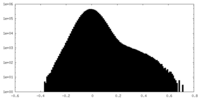
| Density Histograms |
-Half map: #1
| File | emd_35838_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
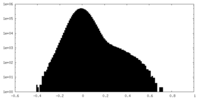
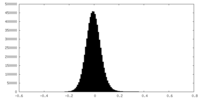
| Density Histograms |
- Sample components
Sample components
-Entire : LysoPS-bound GPR174 in complex with Gs and Nb35.
| Entire | Name: LysoPS-bound GPR174 in complex with Gs and Nb35. |
|---|---|
| Components |
|
-Supramolecule #1: LysoPS-bound GPR174 in complex with Gs and Nb35.
| Supramolecule | Name: LysoPS-bound GPR174 in complex with Gs and Nb35. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.048932 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHVNSGIF ETKFQVDKVN FHMFDVGAQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGCLGNSKTE DQRNEEKAQR EANKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHVNSGIF ETKFQVDKVN FHMFDVGAQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCS VDTENARRIF NDCRDIIQRM HL RQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.845559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody35
| Macromolecule | Name: Nanobody35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #5: Probable G-protein coupled receptor 174
| Macromolecule | Name: Probable G-protein coupled receptor 174 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.90818 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDHHH HHHHHGQPGN GSAFLLAPNG SHAPDHNVTQ QRDEGGSGQP GNGSAFLLAP NGSHAPDHNV TQQRDEENLY FQGVDMPAN YTCTRPDGDN TDFRYFIYAV TYTVILVPGL IGNILALWVF YGYMKETKRA VIFMINLAIA DLLQVLSLPL R IFYYLNHD ...String: DYKDDDDHHH HHHHHGQPGN GSAFLLAPNG SHAPDHNVTQ QRDEGGSGQP GNGSAFLLAP NGSHAPDHNV TQQRDEENLY FQGVDMPAN YTCTRPDGDN TDFRYFIYAV TYTVILVPGL IGNILALWVF YGYMKETKRA VIFMINLAIA DLLQVLSLPL R IFYYLNHD WPFGPGLCMF CFYLKYVNMY ASIYFLVCIS VRRFWFLMYP FRFHDCKQKY DLYISIAGWL IICLACVLFP LL RTSDDTS GNRTKCFVDL PTRNVNLAQS VVMMTIGELI GFVTPLLIVL YCTWKTVLSL QDKYPMAQDL GEKQKALKMI LTC AGVFLI CFAPYHFSFP LDFLVKSNEI KSCLARRVIL IFHSVALCLA SLNSCLDPVI YYFSTNEFRR RLSRQDLHMG SSGG GGSGG GGSSGVFTLE DFVGDWEQTA AYNLDQVLEQ GGVSSLLQNL AVSVTPIQRI VRSGENALKI DIHVIIPYEG LSADQ MAQI EEVFKVVYPV DDHHFKVILP YGTLVIDGVT PNMLNYFGRP YEGIAVFDGK KITVTGTLWN GNKIIDERLI TPDGSM LFR VTINS UniProtKB: Probable G-protein coupled receptor 174 |
-Macromolecule #6: [(2R)-2-oxidanyl-3-phosphonooxy-propyl] (Z)-octadec-9-enoate
| Macromolecule | Name: [(2R)-2-oxidanyl-3-phosphonooxy-propyl] (Z)-octadec-9-enoate type: ligand / ID: 6 / Number of copies: 1 / Formula: UBL |
|---|---|
| Molecular weight | Theoretical: 436.52 Da |
| Chemical component information |  ChemComp-UBL: |
-Macromolecule #7: SERINE
| Macromolecule | Name: SERINE / type: ligand / ID: 7 / Number of copies: 1 / Formula: SER |
|---|---|
| Molecular weight | Theoretical: 105.093 Da |
| Chemical component information |  ChemComp-SER: |
-Macromolecule #8: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 8 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.06 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 132808 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)