+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | hSPCA1 in the CaE1P-ADP state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hSPCA1 / MEMBRANE PROTEIN / METAL TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationGolgi calcium ion homeostasis / Golgi calcium ion transport / P-type manganese transporter activity / trans-Golgi network membrane organization / manganese ion transport / intracellular manganese ion homeostasis / cis-Golgi network membrane / P-type Ca2+ transporter / P-type calcium transporter activity / calcium-dependent cell-cell adhesion ...Golgi calcium ion homeostasis / Golgi calcium ion transport / P-type manganese transporter activity / trans-Golgi network membrane organization / manganese ion transport / intracellular manganese ion homeostasis / cis-Golgi network membrane / P-type Ca2+ transporter / P-type calcium transporter activity / calcium-dependent cell-cell adhesion / positive regulation of Golgi to plasma membrane protein transport / Golgi cisterna membrane / Ion transport by P-type ATPases / epidermis development / trans-Golgi network / calcium ion transmembrane transport / intracellular calcium ion homeostasis / calcium ion transport / manganese ion binding / actin cytoskeleton organization / positive regulation of canonical NF-kappaB signal transduction / Golgi membrane / calcium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||
 Authors Authors | Liu ZM / Wu MQ / Wu C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structure and transport mechanism of the human calcium pump SPCA1. Authors: Mengqi Wu / Cang Wu / Tiefeng Song / Kewu Pan / Yong Wang / Zhongmin Liu /  Abstract: Secretory-pathway Ca-ATPases (SPCAs) play critical roles in maintaining Ca homeostasis, but the exact mechanism of SPCAs-mediated Ca transport remains unclear. Here, we determined six cryo-electron ...Secretory-pathway Ca-ATPases (SPCAs) play critical roles in maintaining Ca homeostasis, but the exact mechanism of SPCAs-mediated Ca transport remains unclear. Here, we determined six cryo-electron microscopy (cryo-EM) structures of human SPCA1 (hSPCA1) in a series of intermediate states, revealing a near-complete conformational cycle. With the aid of molecular dynamics simulations, these structures offer a clear structural basis for Ca entry and release in hSPCA1. We found that hSPCA1 undergoes unique conformational changes during ATP binding and phosphorylation compared to other well-studied P-type II ATPases. In addition, we observed a conformational distortion of the Ca-binding site induced by the separation of transmembrane helices 4L and 6, unveiling a distinct Ca release mechanism. Particularly, we determined a structure of the long-sought CaE2P state of P-type IIA ATPases, providing valuable insights into the Ca transport cycle. Together, these findings enhance our understanding of Ca transport by hSPCA1 and broaden our knowledge of P-type ATPases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35781.map.gz emd_35781.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35781-v30.xml emd-35781-v30.xml emd-35781.xml emd-35781.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35781_fsc.xml emd_35781_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35781.png emd_35781.png | 43 KB | ||
| Filedesc metadata |  emd-35781.cif.gz emd-35781.cif.gz | 6.3 KB | ||
| Others |  emd_35781_half_map_1.map.gz emd_35781_half_map_1.map.gz emd_35781_half_map_2.map.gz emd_35781_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35781 http://ftp.pdbj.org/pub/emdb/structures/EMD-35781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35781 | HTTPS FTP |
-Validation report
| Summary document |  emd_35781_validation.pdf.gz emd_35781_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35781_full_validation.pdf.gz emd_35781_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35781_validation.xml.gz emd_35781_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_35781_validation.cif.gz emd_35781_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35781 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35781 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35781 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35781 | HTTPS FTP |
-Related structure data
| Related structure data |  8iwwMC  8iwpC  8iwrC  8iwsC  8iwtC  8iwuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35781.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35781.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
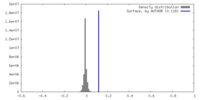
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35781_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35781_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Secretory pathway Ca(2+)-transporting ATPase type 1
| Entire | Name: Secretory pathway Ca(2+)-transporting ATPase type 1 |
|---|---|
| Components |
|
-Supramolecule #1: Secretory pathway Ca(2+)-transporting ATPase type 1
| Supramolecule | Name: Secretory pathway Ca(2+)-transporting ATPase type 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium-transporting ATPase type 2C member 1
| Macromolecule | Name: Calcium-transporting ATPase type 2C member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type Ca2+ transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 97.806797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIPVLTSKKA SELPVSEVAS ILQADLQNGL NKCEVSHRRA FHGWNEFDIS EDEPLWKKYI SQFKNPLIML LLASAVISVL MHQFDDAVS ITVAILIVVT VAFVQEYRSE KSLEELSKLV PPECHCVREG KLEHTLARDL VPGDTVCLSV GDRVPADLRL F EAVDLSID ...String: MIPVLTSKKA SELPVSEVAS ILQADLQNGL NKCEVSHRRA FHGWNEFDIS EDEPLWKKYI SQFKNPLIML LLASAVISVL MHQFDDAVS ITVAILIVVT VAFVQEYRSE KSLEELSKLV PPECHCVREG KLEHTLARDL VPGDTVCLSV GDRVPADLRL F EAVDLSID ESSLTGETTP CSKVTAPQPA ATNGDLASRS NIAFMGTLVR CGKAKGVVIG TGENSEFGEV FKMMQAEEAP KT PLQKSMD LLGKQLSFYS FGIIGIIMLV GWLLGKDILE MFTISVSLAV AAIPEGLPIV VTVTLALGVM RMVKKRAIVK KLP IVETLG CCNVICSDKT GTLTKNEMTV THIFTSDGLH AEVTGVGYNQ FGEVIVDGDV VHGFYNPAVS RIVEAGCVCN DAVI RNNTL MGKPTEGALI ALAMKMGLDG LQQDYIRKAE YPFSSEQKWM AVKCVHRTQQ DRPEICFMKG AYEQVIKYCT TYQSK GQTL TLTQQQRDVY QQEKARMGSA GLRVLALASG PELGQLTFLG LVGIIDPPRT GVKEAVTTLI ASGVSIKMIT GDSQET AVA IASRLGLYSK TSQSVSGEEI DAMDVQQLSQ IVPKVAVFYR ASPRHKMKII KSLQKNGSVV AMTGDGVNDA VALKAAD IG VAMGQTGTDV CKEAADMILV DDDFQTIMSA IEEGKGIYNN IKNFVRFQLS TSIAALTLIS LATLMNFPNP LNAMQILW I NIIMDGPPAQ SLGVEPVDKD VIRKPPRNWK DSILTKNLIL KILVSSIIIV CGTLFVFWRE LRDNVITPRD TTMTFTCFV FFDMFNALSS RSQTKSVFEI GLCSNRMFCY AVLGSIMGQL LVIYFPPLQK VFQTESLSIL DLLFLLGLTS SVCIVAEIIK KVERSREKI QKHV UniProtKB: Calcium-transporting ATPase type 2C member 1 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: TETRAFLUOROALUMINATE ION
| Macromolecule | Name: TETRAFLUOROALUMINATE ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ALF |
|---|---|
| Molecular weight | Theoretical: 102.975 Da |
| Chemical component information |  ChemComp-ALF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)