+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | H7N9 HA-2D7 Fab | |||||||||
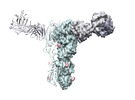
 Map data Map data | The cryoEM of H7N9 HA-2D7 Fab complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | H7N9 / HA / Neutralizing antibody / complex / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  H7N9 subtype (virus) / H7N9 subtype (virus) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhao BB / Sun ZZ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2025 Journal: J Virol / Year: 2025Title: Structural basis of different neutralization capabilities of monoclonal antibodies against H7N9 virus. Authors: Bingbing Zhao / Zhenzhao Sun / Shida Wang / Zhibin Shi / Yongping Jiang / Xiurong Wang / Guohua Deng / Peirong Jiao / Hualan Chen / Jingfei Wang /  Abstract: Neutralizing antibodies (nAbs) are important for the treatment of emerging viral diseases and for effective vaccine development. In this study, we generated and evaluated three nAbs (1H9, 2D7, and ...Neutralizing antibodies (nAbs) are important for the treatment of emerging viral diseases and for effective vaccine development. In this study, we generated and evaluated three nAbs (1H9, 2D7, and C4H4) against H7N9 influenza viruses and found that they differ in their ability to inhibit viral attachment, membrane fusion, and egress. We resolved the cryo-electron microscopy (cryo-EM) structures of H7N9 hemagglutinin (HA) alone and in complex with the nAb antigen-binding fragments (Fabs) and identified the HA head-located epitope for each nAb, thereby revealing the molecular basis and key residues that determine the differences in these nAbs in neutralizing H7N9 viruses. Moreover, we found that the humanized nAb CC4H4 provided complete protection in mice against death caused by a lethal H7N9 virus infection, even when nAb was given 3 days after the mice were infected. These findings provide new insights into the neutralizing mechanism and structural basis for the rational design of H7N9 virus vaccines and therapeutics.IMPORTANCEH7N9 viruses have caused severe infections in both birds and humans since their emergence in early 2013 in China. Their persistent presence and variation in avian populations pose a significant threat to both poultry and humans. There are no treatments for human infections. In this study, we thoroughly investigated the neutralization mechanisms, structural basis, and therapeutic effects of three nAbs (1H9, 2D7, and C4H4) against H7N9 viruses. We revealed the molecular determinants underlying the varied performances of the three nAbs in neutralizing H7N9 viruses and protecting H7N9-infected mice. These insights provide a solid foundation for the rational design of vaccines and therapeutics against H7N9 viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35734.map.gz emd_35734.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35734-v30.xml emd-35734-v30.xml emd-35734.xml emd-35734.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35734.png emd_35734.png | 66.9 KB | ||
| Filedesc metadata |  emd-35734.cif.gz emd-35734.cif.gz | 6.5 KB | ||
| Others |  emd_35734_half_map_1.map.gz emd_35734_half_map_1.map.gz emd_35734_half_map_2.map.gz emd_35734_half_map_2.map.gz | 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35734 http://ftp.pdbj.org/pub/emdb/structures/EMD-35734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35734 | HTTPS FTP |
-Related structure data
| Related structure data |  8iuzMC  8iuxC  8iuyC  8iv0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35734.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35734.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryoEM of H7N9 HA-2D7 Fab complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: The half A map of H7N9 HA-2D7 Fab complex
| File | emd_35734_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half A map of H7N9 HA-2D7 Fab complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The half B map of H7N9 HA-2D7 Fab complex
| File | emd_35734_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half B map of H7N9 HA-2D7 Fab complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The complex of H7N9 HA-1H9 Fab
| Entire | Name: The complex of H7N9 HA-1H9 Fab |
|---|---|
| Components |
|
-Supramolecule #1: The complex of H7N9 HA-1H9 Fab
| Supramolecule | Name: The complex of H7N9 HA-1H9 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  H7N9 subtype (virus) H7N9 subtype (virus) |
-Macromolecule #1: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  H7N9 subtype (virus) H7N9 subtype (virus) |
| Molecular weight | Theoretical: 54.963367 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: DKICLGHHAV SNGTKVNTLT ERGVEVVNAT ETVERTNTPR ICSKGKRTVD LGQCGLLGTI TGPPQCDQFL EFSADLIIER REGSDVCYP GKFVNEEALR QILRESGGID KESMGFTYNG IRTNGVTSAC RRSGSSFYAE MKWLLSNTDN AAFPQMTKSY K NTRESPAI ...String: DKICLGHHAV SNGTKVNTLT ERGVEVVNAT ETVERTNTPR ICSKGKRTVD LGQCGLLGTI TGPPQCDQFL EFSADLIIER REGSDVCYP GKFVNEEALR QILRESGGID KESMGFTYNG IRTNGVTSAC RRSGSSFYAE MKWLLSNTDN AAFPQMTKSY K NTRESPAI IVWGIHHSVS TAEQTKLYGS GNKLVTVGSS NYQQSFVPSP GARPQVNGLS GRIDFHWLIL NPNDTVTFSF NG AFIAPDR ASFLRGKSMG IQSGVQVDAN CEGDCYHSGG TIISNLPFQN IDSRAVGKCP RYVKQRSLLL ATGMKNVPEV PKG KRTARG LFGAIAGFIE NGWEGLIDGW YGFRHQNAQG EGTAADYKST QSAIDQITGK LNRLIAKTNQ QFKLIDNEFN EVEK QIGNV INWTRDSITE VWSYNAELLV AMENQHTIDL ADSEMDKLYE RVKRQLRENA EEDGTGCFEI FHKCDDDCMA SIRNN TYDH RKYREEAMQN UniProtKB: Hemagglutinin |
-Macromolecule #2: 2D7 Fab heavy chain
| Macromolecule | Name: 2D7 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.193651 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVQLQESGPG LVKPSQSLSL TCTVTGYSIT SDYTWNWIRQ FPGNKLEWMG YISYSGSTSY NPSLKSRMSI TRDTSKNQFL LQLNSVTTA DTATYYCTRD GPYWHIDVWG AGTTVTVSS |
-Macromolecule #3: 2D7 Fab light chain
| Macromolecule | Name: 2D7 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.557938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMNQSPSS LSASLGDTIT ITCHASQTIN IWLSWYQLKP GNIPKLLIYK ASNLHTGVPS RFSGSGSGTG FTLTISSLQP EDIASYYCQ QGQSFPYTFG GGTKLEIK |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)