+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | respiratory syncytial virus nucleocapsid-like assembly | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | respiratory syncytial virus / nucleocapsid-like assembly / Cryo-EM / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host PKR/eIFalpha signaling / helical viral capsid / protein serine/threonine kinase inhibitor activity / viral nucleocapsid / host cell cytoplasm / symbiont-mediated suppression of host innate immune response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex / RNA binding Similarity search - Function | |||||||||||||||
| Biological species |  Human respiratory syncytial virus A / Human respiratory syncytial virus A /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||||||||
 Authors Authors | Wang Y / Luo Y / Ling X / Luo B / Jia G / Dong H / Su Z | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Signal Transduct Target Ther / Year: 2023 Journal: Signal Transduct Target Ther / Year: 2023Title: Cryo-EM structure of the nucleocapsid-like assembly of respiratory syncytial virus. Authors: Yan Wang / Chong Zhang / Yongbo Luo / Xiaobin Ling / Bingnan Luo / Guowen Jia / Dan Su / Haohao Dong / Zhaoming Su /  Abstract: Respiratory syncytial virus (RSV) is a nonsegmented, negative strand RNA virus that has caused severe lower respiratory tract infections of high mortality rates in infants and the elderly, yet no ...Respiratory syncytial virus (RSV) is a nonsegmented, negative strand RNA virus that has caused severe lower respiratory tract infections of high mortality rates in infants and the elderly, yet no effective vaccine or antiviral therapy is available. The RSV genome encodes the nucleoprotein (N) that forms helical assembly to encapsulate and protect the RNA genome from degradation, and to serve as a template for transcription and replication. Previous crystal structure revealed a decameric ring architecture of N in complex with the cellular RNA (N-RNA) of 70 nucleotides (70-nt), whereas cryo-ET reconstruction revealed a low-resolution left-handed filament, in which the crystal monomer structure was docked with the helical symmetry applied to simulate a nucleocapsid-like assembly of RSV. However, the molecular details of RSV nucleocapsid assembly remain unknown, which continue to limit our complete understanding of the critical interactions involved in the nucleocapsid and antiviral development that may target this essential process during the viral life cycle. Here we resolve the near-atomic cryo-EM structure of RSV N-RNA that represents roughly one turn of the helical assembly that unveils critical interaction interfaces of RSV nucleocapsid and may facilitate development of RSV antiviral therapy. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35728.map.gz emd_35728.map.gz | 306 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35728-v30.xml emd-35728-v30.xml emd-35728.xml emd-35728.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35728.png emd_35728.png | 28.5 KB | ||
| Filedesc metadata |  emd-35728.cif.gz emd-35728.cif.gz | 6 KB | ||
| Others |  emd_35728_half_map_1.map.gz emd_35728_half_map_1.map.gz emd_35728_half_map_2.map.gz emd_35728_half_map_2.map.gz | 301.7 MB 301.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35728 http://ftp.pdbj.org/pub/emdb/structures/EMD-35728 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35728 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35728 | HTTPS FTP |
-Validation report
| Summary document |  emd_35728_validation.pdf.gz emd_35728_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35728_full_validation.pdf.gz emd_35728_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_35728_validation.xml.gz emd_35728_validation.xml.gz | 16.8 KB | Display | |
| Data in CIF |  emd_35728_validation.cif.gz emd_35728_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35728 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35728 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35728 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35728 | HTTPS FTP |
-Related structure data
| Related structure data |  8iuoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35728.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35728.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35728_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
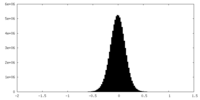
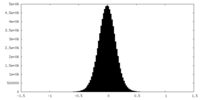
| Density Histograms |
-Half map: #1
| File | emd_35728_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
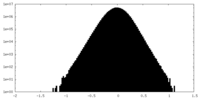
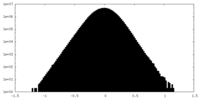
| Density Histograms |
- Sample components
Sample components
-Entire : respiratory syncytial virus nucleocapsid-like assembly
| Entire | Name: respiratory syncytial virus nucleocapsid-like assembly |
|---|---|
| Components |
|
-Supramolecule #1: respiratory syncytial virus nucleocapsid-like assembly
| Supramolecule | Name: respiratory syncytial virus nucleocapsid-like assembly type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus A Human respiratory syncytial virus A |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus A Human respiratory syncytial virus A |
| Molecular weight | Theoretical: 40.198215 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALSKVKLND TLNKDQLLSS SKYTIQRSTG DSIDTPNYDV QKHINKLCGM LLITEDANHK FTGLIGMLYA MSRLGREDTI KILRDAGYH VKANGVDVTT HRQDINGKEM KFEVLTLASL TTEIQINIEI ESRKSYKKML KEMGEVAPEY RHDSPDCGMI I LCIAALVI ...String: MALSKVKLND TLNKDQLLSS SKYTIQRSTG DSIDTPNYDV QKHINKLCGM LLITEDANHK FTGLIGMLYA MSRLGREDTI KILRDAGYH VKANGVDVTT HRQDINGKEM KFEVLTLASL TTEIQINIEI ESRKSYKKML KEMGEVAPEY RHDSPDCGMI I LCIAALVI TKLAAGDRSG LTAVIRRANN VLKNEMKRYK GLLPKDIANS FYEVFEKHPH FIDVFVHFGI AQSSTRGGSR VE GIFAGLF MNAYGAGQVM LRWGVLAKSV KNIMLGHASV QAEMEQVVEV YEYAQKLGGE AGFYHILNNP KASLLSLTQF PHF SSVVLG NAAGLGIMGE YRGTPRNQDL YDAAKAYAEQ LKENGV UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (35-MER)
| Macromolecule | Name: RNA (35-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.670844 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)