[English] 日本語
 Yorodumi
Yorodumi- EMDB-35727: Cryo-EM structure of the CRT-LESS RC-LH core complex from roseifl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CRT-LESS RC-LH core complex from roseiflexus castenholzii | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CRT-LESS RC-LH CORE COMPLEX / ROSEIFLEXUS CASTENHOLZII / PHOTOSYNTHESIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / photosynthesis, light reaction / endomembrane system / electron transfer activity / iron ion binding / heme binding / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Roseiflexus castenholzii (bacteria) Roseiflexus castenholzii (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | ||||||||||||
 Authors Authors | Wang G-L / Qi C-H / Yu L-J | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes. Authors: Chen-Hui Qi / Guang-Lei Wang / Fang-Fang Wang / Yueyong Xin / Mei-Juan Zou / Michael T Madigan / Zheng-Yu Wang-Otomo / Fei Ma / Long-Jiang Yu /    Abstract: In wild-type phototrophic organisms, carotenoids (Crts) are primarily packed into specific pigment-protein complexes along with (Bacterio)chlorophylls and play important roles in the photosynthesis. ...In wild-type phototrophic organisms, carotenoids (Crts) are primarily packed into specific pigment-protein complexes along with (Bacterio)chlorophylls and play important roles in the photosynthesis. Diphenylamine (DPA) inhibits carotenogenesis but not phototrophic growth of anoxygenic phototrophs and eliminates virtually all Crts from photocomplexes. To investigate the effect of Crts on assembly of the reaction center-light-harvesting (RC-LH) complex from the filamentous anoxygenic phototroph Roseiflexus (Rfl.) castenholzii, we generated carotenoidless (Crt-less) RC-LH complexes by growing cells in the presence of DPA. Here, we present cryo-EM structures of the Rfl. castenholzii native and Crt-less RC-LH complexes with resolutions of 2.86 Å and 2.85 Å, respectively. From the high-quality map obtained, several important but previously unresolved details in the Rfl. castenholzii RC-LH structure were determined unambiguously including the assignment and likely function of three small polypeptides, and the content and spatial arrangement of Crts with bacteriochlorophyll molecules. The overall structures of Crt-containing and Crt-less complexes are similar. However, structural comparisons showed that only five Crts remain in complexes from DPA-treated cells and that the subunit X (TMx) flanked on the N-terminal helix of the Cyt-subunit is missing. Based on these results, the function of Crts in the assembly of the Rfl. castenholzii RC-LH complex and the molecular mechanism of quinone exchange is discussed. These structural details provide a fresh look at the photosynthetic apparatus of an evolutionary ancient phototroph as well as new insights into the importance of Crts for proper assembly and functioning of the RC-LH complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35727.map.gz emd_35727.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35727-v30.xml emd-35727-v30.xml emd-35727.xml emd-35727.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35727_fsc.xml emd_35727_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_35727.png emd_35727.png | 62.9 KB | ||
| Filedesc metadata |  emd-35727.cif.gz emd-35727.cif.gz | 7.8 KB | ||
| Others |  emd_35727_half_map_1.map.gz emd_35727_half_map_1.map.gz emd_35727_half_map_2.map.gz emd_35727_half_map_2.map.gz | 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35727 http://ftp.pdbj.org/pub/emdb/structures/EMD-35727 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35727 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35727 | HTTPS FTP |
-Related structure data
| Related structure data |  8iunMC  8iugC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35727.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35727.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
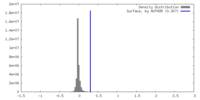
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35727_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
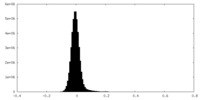
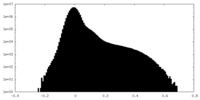
| Density Histograms |
-Half map: #1
| File | emd_35727_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
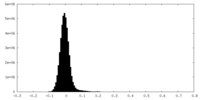
| Density Histograms |
- Sample components
Sample components
+Entire : CRT-LESS RC-LH CORE COMPLEX
+Supramolecule #1: CRT-LESS RC-LH CORE COMPLEX
+Macromolecule #1: Antenna complex alpha/beta subunit
+Macromolecule #2: Reaction center protein L chain
+Macromolecule #3: Alpha subunit of light-harvesting 1
+Macromolecule #4: reaction center small polypeptide
+Macromolecule #5: reaction center small polypeptide
+Macromolecule #6: Cytochrome subunit of photosynthetic reaction center
+Macromolecule #7: reaction center unknown polypeptide
+Macromolecule #8: BACTERIOCHLOROPHYLL A
+Macromolecule #9: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #10: gamma-Carotene
+Macromolecule #11: BACTERIOPHEOPHYTIN A
+Macromolecule #12: MANGANESE (II) ION
+Macromolecule #13: 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,...
+Macromolecule #14: 2-O-octyl-beta-D-glucopyranose
+Macromolecule #15: DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE
+Macromolecule #16: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #17: UNKNOWN LIGAND
+Macromolecule #18: CARDIOLIPIN
+Macromolecule #19: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #20: HEME C
+Macromolecule #21: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)