[English] 日本語
 Yorodumi
Yorodumi- EMDB-35657: Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha rece... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Complex / Prostaglandin receptor / Agonist / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprostaglandin F receptor activity / parturition / Prostanoid ligand receptors / cellular response to prostaglandin D stimulus / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress ...prostaglandin F receptor activity / parturition / Prostanoid ligand receptors / cellular response to prostaglandin D stimulus / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / response to estradiol / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / response to lipopolysaccharide / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / GTPase activity / positive regulation of cell population proliferation / synapse / positive regulation of gene expression / negative regulation of apoptotic process / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / extracellular region / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Lv X / Gao K / Nie J / Zhang X / Zhang S / Ren Y / Li Q / Huang J / Liu L / Sun X ...Lv X / Gao K / Nie J / Zhang X / Zhang S / Ren Y / Li Q / Huang J / Liu L / Sun X / Zhang W / Liu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structures of human prostaglandin F receptor reveal the mechanism of ligand and G protein selectivity. Authors: Xiuqing Lv / Kaixuan Gao / Jia Nie / Xin Zhang / Shuhao Zhang / Yinhang Ren / Xiaoou Sun / Qi Li / Jingrui Huang / Lijuan Liu / Xiaowen Zhang / Weishe Zhang / Xiangyu Liu /  Abstract: Prostaglandins and their receptors regulate various physiological processes. Carboprost, an analog of prostaglandin F and an agonist for the prostaglandin F2-alpha receptor (FP receptor), is ...Prostaglandins and their receptors regulate various physiological processes. Carboprost, an analog of prostaglandin F and an agonist for the prostaglandin F2-alpha receptor (FP receptor), is clinically used to treat postpartum hemorrhage (PPH). However, off-target activation of closely related receptors such as the prostaglandin E receptor subtype EP3 (EP3 receptor) by carboprost results in side effects and limits the clinical application. Meanwhile, the FP receptor selective agonist latanoprost is not suitable to treat PPH due to its poor solubility and fast clearance. Here, we present two cryo-EM structures of the FP receptor bound to carboprost and latanoprost-FA (the free acid form of latanoprost) at 2.7 Å and 3.2 Å resolution, respectively. The structures reveal the molecular mechanism of FP receptor selectivity for both endogenous prostaglandins and clinical drugs, as well as the molecular mechanism of G protein coupling preference by the prostaglandin receptors. The structural information may guide the development of better prostaglandin drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35657.map.gz emd_35657.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35657-v30.xml emd-35657-v30.xml emd-35657.xml emd-35657.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35657.png emd_35657.png | 46.8 KB | ||
| Masks |  emd_35657_msk_1.map emd_35657_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35657.cif.gz emd-35657.cif.gz | 6.7 KB | ||
| Others |  emd_35657_half_map_1.map.gz emd_35657_half_map_1.map.gz emd_35657_half_map_2.map.gz emd_35657_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35657 http://ftp.pdbj.org/pub/emdb/structures/EMD-35657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35657 | HTTPS FTP |
-Validation report
| Summary document |  emd_35657_validation.pdf.gz emd_35657_validation.pdf.gz | 783.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35657_full_validation.pdf.gz emd_35657_full_validation.pdf.gz | 782.7 KB | Display | |
| Data in XML |  emd_35657_validation.xml.gz emd_35657_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_35657_validation.cif.gz emd_35657_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35657 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35657 | HTTPS FTP |
-Related structure data
| Related structure data |  8iq4MC  8iq6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35657.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35657.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
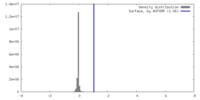
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35657_msk_1.map emd_35657_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
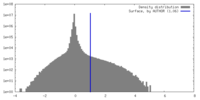
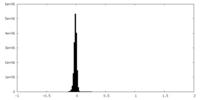
| Density Histograms |
-Half map: #1
| File | emd_35657_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
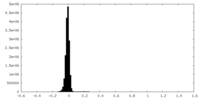
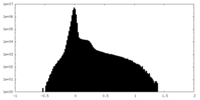
| Density Histograms |
-Half map: #2
| File | emd_35657_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
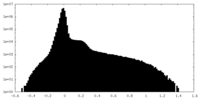
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha rece...
| Entire | Name: Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha rece...
| Supramolecule | Name: Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Prostaglandin F2-alpha receptor
| Macromolecule | Name: Prostaglandin F2-alpha receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.909363 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAMS MNNSKQLVSP AAALLSNTTC QTENRLSVFF SVIFMTVGIL SNSLAIAILM KAYQRFRQKS KASFLLLASG LVITDFFGH LINGAIAVFV YASDKEWIRF DQSNVLCSIF GICMVFSGLC PLLLGSVMAI ERCIGVTKPI FHSTKITSKH V KMMLSGVC ...String: DYKDDDDAMS MNNSKQLVSP AAALLSNTTC QTENRLSVFF SVIFMTVGIL SNSLAIAILM KAYQRFRQKS KASFLLLASG LVITDFFGH LINGAIAVFV YASDKEWIRF DQSNVLCSIF GICMVFSGLC PLLLGSVMAI ERCIGVTKPI FHSTKITSKH V KMMLSGVC LFAVFIALLP ILGHRDYKIQ ASRTWCFYNT EDIKDWEDRF YLLLFSFLGL LALGVSLLCN AITGITLLRV KF KSQQHRQ GRSHHLEMVI QLLAIMCVSC ICWSPFLVTM ANIGINGNHS LETCETTLFA LRMATWNQIL DPWVYILLRK AVL KNLYKL ASQCCGVHVI SLHIWELSSI KNSLKVAAIS ESPVAEKSAS THHHHHHGGS GGLEVLFQ UniProtKB: Prostaglandin F2-alpha receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.168926 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR ...String: GPTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR YTTPEDATPE PGEDPRVTRA KYFIRDEFLR ISTASGDGRH YCYPHFTCAV DTENARRIFN DCKDIILQMN LR EYNLV UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.686328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTPFCFD VTSTTYAYRG QGTQVTVSSH HHHHH |
-Macromolecule #6: Z-7-[(1R,2R,3R,5S)-2-[(E,3S)-3-methyl-3-oxidanyl-oct-1-enyl]-3,5-...
| Macromolecule | Name: Z-7-[(1R,2R,3R,5S)-2-[(E,3S)-3-methyl-3-oxidanyl-oct-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: 87Q |
|---|---|
| Molecular weight | Theoretical: 368.508 Da |
| Chemical component information | 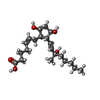 ChemComp-87Q: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.45 |
|---|---|
| Grid | Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 327293 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)