+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herg1a-herg1b closed state 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | channelrhodopsin / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated potassium channel activity / monoatomic ion channel complex / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhang MF | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of Herg1a-herg1b closed state 1 Authors: Zhang MF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35614.map.gz emd_35614.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35614-v30.xml emd-35614-v30.xml emd-35614.xml emd-35614.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35614.png emd_35614.png | 47 KB | ||
| Filedesc metadata |  emd-35614.cif.gz emd-35614.cif.gz | 5.9 KB | ||
| Others |  emd_35614_half_map_1.map.gz emd_35614_half_map_1.map.gz emd_35614_half_map_2.map.gz emd_35614_half_map_2.map.gz | 225.9 MB 225.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35614 http://ftp.pdbj.org/pub/emdb/structures/EMD-35614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35614 | HTTPS FTP |
-Validation report
| Summary document |  emd_35614_validation.pdf.gz emd_35614_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35614_full_validation.pdf.gz emd_35614_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35614_validation.xml.gz emd_35614_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_35614_validation.cif.gz emd_35614_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35614 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35614 | HTTPS FTP |
-Related structure data
| Related structure data |  8iobMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35614.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35614.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.849 Å | ||||||||||||||||||||||||||||||||||||
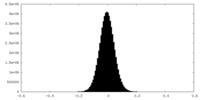
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35614_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
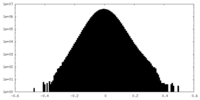
| Projections & Slices |
| ||||||||||||
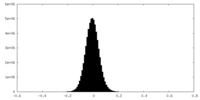
| Density Histograms |
-Half map: #2
| File | emd_35614_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
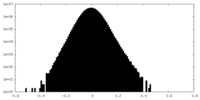
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Herg1a-herg1b channel
| Entire | Name: Herg1a-herg1b channel |
|---|---|
| Components |
|
-Supramolecule #1: Herg1a-herg1b channel
| Supramolecule | Name: Herg1a-herg1b channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium voltage-gated channel subfamily H member 2
| Macromolecule | Name: Potassium voltage-gated channel subfamily H member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 126.802727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SPAHDTNHRG PPTSWLAPGR A KTFRLKLP ...String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SPAHDTNHRG PPTSWLAPGR A KTFRLKLP ALLALTARES SVRSGGAGGA GAPGAVVVDV DLTPAAPSSE SLALDEVTAM DNHVAGLGPA EERRALVGPG SP PRSAPGQ LPSPRAHSLN PDASGSSCSL ARTRSRESCA SVRRASSADD IEAMRAGVLP PPPRHASTGA MHPLRSGLLN STS DSDLVR YRTISKIPQI TLNFVDLKGD PFLASPTSDR EIIAPKIKER THNVTEKVTQ VLSLGADVLP EYKLQAPRIH RWTI LHYSP FKAVWDWLIL LLVIYTAVFT PYSAAFLLKE TEEGPPATEC GYACQPLAVV DLIVDIMFIV DILINFRTTY VNANE EVVS HPGRIAVHYF KGWFLIDMVA AIPFDLLIFG SGSEELIGLL KTARLLRLVR VARKLDRYSE YGAAVLFLLM CTFALI AHW LACIWYAIGN MEQPHMDSRI GWLHNLGDQI GKPYNSSGLG GPSIKDKYVT ALYFTFSSLT SVGFGNVSPN TNSEKIF SI CVMLIGSLMY ASIFGNVSAI IQRLYSGTAR YHTQMLRVRE FIRFHQIPNP LRQRLEEYFQ HAWSYTNGID MNAVLKGF P ECLQADICLH LNRSLLQHCK PFRGATKGCL RALAMKFKTT HAPPGDTLVH AGDLLTALYF ISRGSIEILR GDVVVAILG KNDIFGEPLN LYARPGKSNG DVRALTYCDL HKIHRDDLLE VLDMYPEFSD HFWSSLEITF NLRDTNMIPG SPGSTELEGG FSRQRKRKL SFRRRTDKDT EQPGEVSALG PGRAGAGPSS RGRPGGPWGE SPSSGPSSPE SSEDEGPGRS SSPLRLVPFS S PRPPGEPP GGEPLMEDCE KSSDTCNPLS GAFSGVSNIF SFWGDSRGRQ YQELPRCPAP TPSLLNIPLS SPGRRPRGDV ES RLDALQR QLNRLETRLS ADMATVLQLL QRQMTLVPPA YSAVTTPGPG PTSTSPLLPV SPLPTLTLDS LSQVSQFMAC EEL PPGAPE LPQEGPTRRL SLPGQLGALT SQPLHRHGSD PGS UniProtKB: Voltage-gated inwardly rectifying potassium channel KCNH2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)