[English] 日本語
 Yorodumi
Yorodumi- EMDB-3558: Centriolar Proximal MT triplet in centrosomes extracted from o.ar... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3558 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Centriolar Proximal MT triplet in centrosomes extracted from o.aries thymocytes | |||||||||
 Map data Map data | Centriolar proximal MT triplet from a mammal centrosome. Centrosomes were extracted from o.aries thymocytes. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
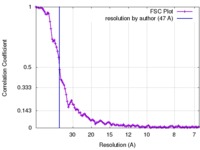
| Method | subtomogram averaging / cryo EM / Resolution: 47.0 Å | |||||||||
 Authors Authors | Busselez J / Chichon FJ / Melero R / Carrascosa JL / Carazo JM | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2019 Journal: Sci Rep / Year: 2019Title: Cryo-Electron Tomography and Proteomics studies of centrosomes from differentiated quiescent thymocytes. Authors: Johan Busselez / Francisco Javier Chichón / Maria Josefa Rodríguez / Adan Alpízar / Séverine Isabelle Gharbi / Mònica Franch / Roberto Melero / Alberto Paradela / José L Carrascosa / José-Maria Carazo /   Abstract: We have used cryo Electron Tomography, proteomics and immunolabeling to study centrosomes isolated from the young lamb thymus, an efficient source of quiescent differentiated cells. We compared the ...We have used cryo Electron Tomography, proteomics and immunolabeling to study centrosomes isolated from the young lamb thymus, an efficient source of quiescent differentiated cells. We compared the proteome of thymocyte centrosomes to data published for KE37 cells, focusing on proteins associated with centriole disengagement and centrosome separation. The data obtained enhances our understanding of the protein system joining the centrioles, a system comprised of a branched network of fibers linked to an apparently amorphous density that was partially characterized here. A number of proteins were localized to the amorphous density by immunolabeling (C-NAP1, cohesin SMC1, condensin SMC4 and NCAPD2), yet not DNA. In conjuction, these data not only extend our understanding of centrosomes but they will help refine the model that focus on the protein system associated with the centriolar junction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3558.map.gz emd_3558.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3558-v30.xml emd-3558-v30.xml emd-3558.xml emd-3558.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3558_fsc.xml emd_3558_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_3558.png emd_3558.png | 40.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3558 http://ftp.pdbj.org/pub/emdb/structures/EMD-3558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3558 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Centriolar proximal MT triplet from a mammal centrosome. Centrosomes were extracted from o.aries thymocytes. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Centrosome enriched from young lamb thymocytes
| Entire | Name: Centrosome enriched from young lamb thymocytes |
|---|---|
| Components |
|
-Supramolecule #1: Centrosome enriched from young lamb thymocytes
| Supramolecule | Name: Centrosome enriched from young lamb thymocytes / type: organelle_or_cellular_component / ID: 1 / Parent: 0 Details: The centrosome were extracted accordingly to Komesli et al. 1989 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 230000 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.076 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Component - Concentration: 10.0 mM / Component - Formula: C8H18N2O6S2 / Component - Name: Pipes / Details: pH rectification with KOH |
| Grid | Model: quantifoil 3.5/1 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: FORMVAR / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
| Details | The sample was enriched by two step of sedimentation in sucrose gradient. The sucrose was washed out in several drops of buffer, the last of which included fiducial markers (10 nm bovine serum albumin (BSA)-coated gold beads) for tilt series alignment |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: Gatan 968 Quantum / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average exposure time: 1.0 sec. / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 5.906 µm / Calibrated defocus min: 5.54 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)