+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Filament interface structure of GAC with phosphate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glutaminase / phosphate / filament / PROTEIN FIBRIL / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-glutamine catabolic process / regulation of respiratory gaseous exchange by nervous system process / glutamate biosynthetic process / Glutamate and glutamine metabolism / glutaminase / intracellular glutamate homeostasis / Glutamate Neurotransmitter Release Cycle / glutaminase activity / suckling behavior / TP53 Regulates Metabolic Genes ...L-glutamine catabolic process / regulation of respiratory gaseous exchange by nervous system process / glutamate biosynthetic process / Glutamate and glutamine metabolism / glutaminase / intracellular glutamate homeostasis / Glutamate Neurotransmitter Release Cycle / glutaminase activity / suckling behavior / TP53 Regulates Metabolic Genes / protein homotetramerization / chemical synaptic transmission / mitochondrial matrix / synapse / mitochondrion / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
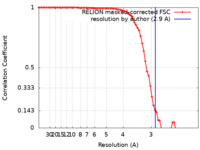
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Guo CJ / Liu JL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Structural basis for activation and filamentation of glutaminase. Authors: Chen-Jun Guo / Zi-Xuan Wang / Ji-Long Liu /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35574.map.gz emd_35574.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35574-v30.xml emd-35574-v30.xml emd-35574.xml emd-35574.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35574_fsc.xml emd_35574_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_35574.png emd_35574.png | 45.1 KB | ||
| Masks |  emd_35574_msk_1.map emd_35574_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35574.cif.gz emd-35574.cif.gz | 5.4 KB | ||
| Others |  emd_35574_half_map_1.map.gz emd_35574_half_map_1.map.gz emd_35574_half_map_2.map.gz emd_35574_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35574 http://ftp.pdbj.org/pub/emdb/structures/EMD-35574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35574 | HTTPS FTP |
-Validation report
| Summary document |  emd_35574_validation.pdf.gz emd_35574_validation.pdf.gz | 717 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35574_full_validation.pdf.gz emd_35574_full_validation.pdf.gz | 716.5 KB | Display | |
| Data in XML |  emd_35574_validation.xml.gz emd_35574_validation.xml.gz | 11.9 KB | Display | |
| Data in CIF |  emd_35574_validation.cif.gz emd_35574_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35574 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35574 | HTTPS FTP |
-Related structure data
| Related structure data |  8imbMC  8imaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35574.map.gz / Format: CCP4 / Size: 5.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35574.map.gz / Format: CCP4 / Size: 5.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35574_msk_1.map emd_35574_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35574_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35574_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GAC filament with phosphate
| Entire | Name: GAC filament with phosphate |
|---|---|
| Components |
|
-Supramolecule #1: GAC filament with phosphate
| Supramolecule | Name: GAC filament with phosphate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glutaminase kidney isoform, mitochondrial
| Macromolecule | Name: Glutaminase kidney isoform, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glutaminase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.083738 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LVASGENKIK QGLLPSLEDL LFYTIAEGQE KIPVHKFITA LKSTGLRTSD PRLKECMDML RLTLQTTSDG VMLDKDLFKK CVQSNIVLL TQAFRRKFVI PDFMSFTSHI DELYESAKKQ SGGKVADYIP QLAKFSPDLW GVSVCTVDGQ RHSTGDTKVP F CLQSCVKP ...String: LVASGENKIK QGLLPSLEDL LFYTIAEGQE KIPVHKFITA LKSTGLRTSD PRLKECMDML RLTLQTTSDG VMLDKDLFKK CVQSNIVLL TQAFRRKFVI PDFMSFTSHI DELYESAKKQ SGGKVADYIP QLAKFSPDLW GVSVCTVDGQ RHSTGDTKVP F CLQSCVKP LKYAIAVNDL GTEYVHRYVG KEPSGLRFNK LFLNEDDKPH NPMVNAGAIV VTSLIKQGVN NAEKFDYVMQ FL NKMAGNE YVGFSNATFQ SERESGDRNF AIGYYLKEKK CFPEGTDMVG ILDFYFQLCS IEVTCESASV MAATLANGGF CPI TGERVL SPEAVRNTLS LMHSCGMYDF SGQFAFHVGL PAKSGVAGGI LLVVPNVMGM MCWSPPLDKM GNSVKGIHFC HDLV SLCNF HNYDNLRHFA KKLDPRREGG DQRHSFGPLD YESLQQELAL KETVWKKVSP ESNEDISTTV VYRMESLGEK S UniProtKB: Glutaminase kidney isoform, mitochondrial |
-Macromolecule #2: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)