[English] 日本語
 Yorodumi
Yorodumi- EMDB-35499: Complex structure of Neuropeptide Y Y2 receptor in complex with P... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi (Focused map on Gi-scFv16) | |||||||||
 Map data Map data | Focused map (Gi-scFv16) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
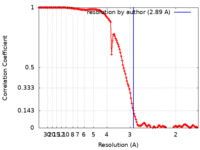
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Kang H / Park C / Kim J / Choi H-J | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling. Authors: Hyunook Kang / Chaehee Park / Yeol Kyo Choi / Jungnam Bae / Sohee Kwon / Jinuk Kim / Chulwon Choi / Chaok Seok / Wonpil Im / Hee-Jung Choi /   Abstract: Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors ...Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors (YR and YR, respectively) activate G signaling, but their physiological responses to food intake are different. In addition, deletion of the two N-terminal amino acids of peptide YY (PYY(3-36)), the endogenous form found in circulation, can stimulate YR but not YR, suggesting that YR and YR may have distinct ligand-binding modes. Here, we report the cryo-electron microscopy structures of the PYY(3-36)‒YR‒G and NPY‒YR‒G complexes. Using cell-based assays, molecular dynamics simulations, and structural analysis, we revealed the molecular basis of the exclusive binding of PYY(3-36) to YR. Furthermore, we demonstrated that YR favors G protein signaling over β-arrestin signaling upon activation, whereas YR does not show a preference between these two pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35499.map.gz emd_35499.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35499-v30.xml emd-35499-v30.xml emd-35499.xml emd-35499.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35499_fsc.xml emd_35499_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35499.png emd_35499.png | 30.4 KB | ||
| Others |  emd_35499_half_map_1.map.gz emd_35499_half_map_1.map.gz emd_35499_half_map_2.map.gz emd_35499_half_map_2.map.gz | 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35499 http://ftp.pdbj.org/pub/emdb/structures/EMD-35499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35499 | HTTPS FTP |
-Validation report
| Summary document |  emd_35499_validation.pdf.gz emd_35499_validation.pdf.gz | 948.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35499_full_validation.pdf.gz emd_35499_full_validation.pdf.gz | 948.4 KB | Display | |
| Data in XML |  emd_35499_validation.xml.gz emd_35499_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_35499_validation.cif.gz emd_35499_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35499 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35499 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35499.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35499.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map (Gi-scFv16) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.849 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B
| File | emd_35499_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
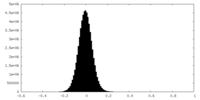
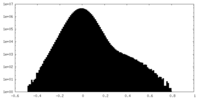
| Density Histograms |
-Half map: Half map A
| File | emd_35499_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
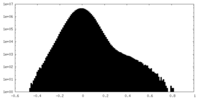
| Density Histograms |
- Sample components
Sample components
-Entire : Complex structure of PYY(3-36)-Y2R-Gi-scFv16
| Entire | Name: Complex structure of PYY(3-36)-Y2R-Gi-scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: Complex structure of PYY(3-36)-Y2R-Gi-scFv16
| Supramolecule | Name: Complex structure of PYY(3-36)-Y2R-Gi-scFv16 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Guanine nucleotide-binding protein
| Supramolecule | Name: Guanine nucleotide-binding protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3, #5 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: scFv16
| Supramolecule | Name: scFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #6 |
|---|
-Supramolecule #4: PYY(3-36)
| Supramolecule | Name: PYY(3-36) / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #4 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)