+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Potassium voltage-gated channel subfamily KQT member 2 / Ebio1 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaxon initial segment / Voltage gated Potassium channels / node of Ranvier / Interaction between L1 and Ankyrins / ankyrin binding / CaM pathway / Cam-PDE 1 activation / voltage-gated monoatomic cation channel activity / Sodium/Calcium exchangers / Calmodulin induced events ...axon initial segment / Voltage gated Potassium channels / node of Ranvier / Interaction between L1 and Ankyrins / ankyrin binding / CaM pathway / Cam-PDE 1 activation / voltage-gated monoatomic cation channel activity / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / Loss of phosphorylation of MECP2 at T308 / CREB1 phosphorylation through the activation of Adenylate Cyclase / negative regulation of high voltage-gated calcium channel activity / PKA activation / CaMK IV-mediated phosphorylation of CREB / Glycogen breakdown (glycogenolysis) / CLEC7A (Dectin-1) induces NFAT activation / Activation of RAC1 downstream of NMDARs / negative regulation of ryanodine-sensitive calcium-release channel activity / organelle localization by membrane tethering / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential / presynaptic endocytosis / regulation of cell communication by electrical coupling involved in cardiac conduction / Synthesis of IP3 and IP4 in the cytosol / Phase 0 - rapid depolarisation / Negative regulation of NMDA receptor-mediated neuronal transmission / calcineurin-mediated signaling / Unblocking of NMDA receptors, glutamate binding and activation / RHO GTPases activate PAKs / regulation of ryanodine-sensitive calcium-release channel activity / Ion transport by P-type ATPases / Uptake and function of anthrax toxins / Long-term potentiation / protein phosphatase activator activity / action potential / Calcineurin activates NFAT / voltage-gated potassium channel activity / Regulation of MECP2 expression and activity / DARPP-32 events / Smooth Muscle Contraction / detection of calcium ion / regulation of cardiac muscle contraction / catalytic complex / RHO GTPases activate IQGAPs / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / calcium channel inhibitor activity / Activation of AMPK downstream of NMDARs / presynaptic cytosol / cellular response to interferon-beta / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Protein methylation / Ion homeostasis / eNOS activation / titin binding / Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / regulation of calcium-mediated signaling / voltage-gated potassium channel complex / potassium ion transmembrane transport / FCERI mediated Ca+2 mobilization / calcium channel complex / substantia nigra development / regulation of heart rate / FCGR3A-mediated IL10 synthesis / Ras activation upon Ca2+ influx through NMDA receptor / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / calyx of Held / adenylate cyclase activator activity / protein serine/threonine kinase activator activity / VEGFR2 mediated cell proliferation / sarcomere / regulation of cytokinesis / VEGFR2 mediated vascular permeability / spindle microtubule / positive regulation of receptor signaling pathway via JAK-STAT / Translocation of SLC2A4 (GLUT4) to the plasma membrane / calcium channel regulator activity / RAF activation / Transcriptional activation of mitochondrial biogenesis / response to calcium ion / cellular response to type II interferon / G2/M transition of mitotic cell cycle / Stimuli-sensing channels / spindle pole / Signaling by RAF1 mutants / RAS processing / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / calcium-dependent protein binding / Signaling by BRAF and RAF1 fusions / long-term synaptic potentiation / Platelet degranulation / sperm midpiece / nervous system development Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Ma D / Guo J | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: A small-molecule activation mechanism that directly opens the KCNQ2 channel. Authors: Shaoying Zhang / Demin Ma / Kun Wang / Ya Li / Zhenni Yang / Xiaoxiao Li / Junnan Li / Jiangnan He / Lianghe Mei / Yangliang Ye / Zongsheng Chen / Juwen Shen / Panpan Hou / Jiangtao Guo / ...Authors: Shaoying Zhang / Demin Ma / Kun Wang / Ya Li / Zhenni Yang / Xiaoxiao Li / Junnan Li / Jiangnan He / Lianghe Mei / Yangliang Ye / Zongsheng Chen / Juwen Shen / Panpan Hou / Jiangtao Guo / Qiansen Zhang / Huaiyu Yang /  Abstract: Pharmacological activation of voltage-gated ion channels by ligands serves as the basis for therapy and mainly involves a classic gating mechanism that augments the native voltage-dependent open ...Pharmacological activation of voltage-gated ion channels by ligands serves as the basis for therapy and mainly involves a classic gating mechanism that augments the native voltage-dependent open probability. Through structure-based virtual screening, we identified a new scaffold compound, Ebio1, serving as a potent and subtype-selective activator for the voltage-gated potassium channel KCNQ2 and featuring a new activation mechanism. Single-channel patch-clamp, cryogenic-electron microscopy and molecular dynamic simulations, along with chemical derivatives, reveal that Ebio1 engages the KCNQ2 activation by generating an extended channel gate with a larger conductance at the saturating voltage (+50 mV). This mechanism is different from the previously observed activation mechanism of ligands on voltage-gated ion channels. Ebio1 caused S6 helices from residues S303 and F305 to perform a twist-to-open movement, which was sufficient to open the KCNQ2 gate. Overall, our findings provide mechanistic insights into the activation of KCNQ2 channel by Ebio1 and lend support for KCNQ-related drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35487.map.gz emd_35487.map.gz | 47.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35487-v30.xml emd-35487-v30.xml emd-35487.xml emd-35487.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35487.png emd_35487.png | 58 KB | ||
| Filedesc metadata |  emd-35487.cif.gz emd-35487.cif.gz | 5.9 KB | ||
| Others |  emd_35487_half_map_1.map.gz emd_35487_half_map_1.map.gz emd_35487_half_map_2.map.gz emd_35487_half_map_2.map.gz | 37.1 MB 37.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35487 http://ftp.pdbj.org/pub/emdb/structures/EMD-35487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35487 | HTTPS FTP |
-Related structure data
| Related structure data |  8ijkMC  8x43C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35487.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35487.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
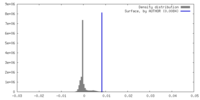
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35487_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35487_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human KCNQ2-CaM-Ebio1 complex in the presence of PIP2
| Entire | Name: human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 |
|---|---|
| Components |
|
-Supramolecule #1: human KCNQ2-CaM-Ebio1 complex in the presence of PIP2
| Supramolecule | Name: human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium voltage-gated channel subfamily KQT member 2
| Macromolecule | Name: Potassium voltage-gated channel subfamily KQT member 2 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.627812 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGKPPKRNA FYRKLQNFLY NVLERPRGWA FIYHAYVFLL VFSCLVLSVF STIKEYEKSS EGALYILEIV TIVVFGVEYF VRIWAAGCC CRYRGWRGRL KFARKPFCVI DIMVLIASIA VLAAGSQGNV FATSALRSLR FLQILRMIRM DRRGGTWKLL G SVVYAHSK ...String: MAGKPPKRNA FYRKLQNFLY NVLERPRGWA FIYHAYVFLL VFSCLVLSVF STIKEYEKSS EGALYILEIV TIVVFGVEYF VRIWAAGCC CRYRGWRGRL KFARKPFCVI DIMVLIASIA VLAAGSQGNV FATSALRSLR FLQILRMIRM DRRGGTWKLL G SVVYAHSK ELVTAWYIGF LCLILASFLV YLAEKGENDH FDTYADALWW GLITLTTIGY GDKYPQTWNG RLLAATFTLI GV SFFALPA GILGSGFALK VQEQHRQKHF EKRRNPAAGL IQSAWRFYAT NLSRTDLHST WQYYERTVTV PMYSSQTQTY GAS RLIPPL NQLELLRNLK SKSGLAFRKD PPPEPSPSKG SPCRGPLCGC CPGRSSQKVS LKDRVFSSPR GVAAKGKGSP QAQT VRRSP SADQSLEDSP SKVPKSWSFG DRSRARQAFR IKGAASRQNS EEASLPGEDI VDDKSCPCEF VTEDLTPGLK VSIRA VCVM RFLVSKRKFK ESLRPYDVMD VIEQYSAGHL DMLSRIKSLQ SRVDQIVGRG PAITDKDRTK GPAEAELPED PSMMGR LGK VEKQVLSMEK KLDFLVNIYM QRMGIPPTET EAYFGAKEPE PAPPYHSPED SREHVDRHGC IVKIVRSSSS TGQKNFS VE GGSSGGWSHP QFEK UniProtKB: Potassium voltage-gated channel subfamily KQT member 2 |
-Macromolecule #2: Calmodulin-1
| Macromolecule | Name: Calmodulin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.852545 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADQLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTIDFPEFL TMMARKMKDT DSEEEIREA FRVFDKDGNG YISAAELRHV MTNLGEKLTD EEVDEMIREA DIDGDGQVNY EEFVQMMTAK UniProtKB: Calmodulin-1 |
-Macromolecule #3: N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide
| Macromolecule | Name: N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide type: ligand / ID: 3 / Number of copies: 4 / Formula: 7PN |
|---|---|
| Molecular weight | Theoretical: 291.319 Da |
| Chemical component information |  ChemComp-7PN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 28232 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)