[English] 日本語
 Yorodumi
Yorodumi- EMDB-35351: Cryo-EM structure of CXCL8 bound C-X-C chemokine receptor 1 in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CXCL8 bound C-X-C chemokine receptor 1 in complex with Gi heterotrimer | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Chemokine / Interleukin / CXCR / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of single stranded viral RNA replication via double stranded DNA intermediate / regulation of entry of bacterium into host cell / interleukin-8 receptor binding / interleukin-8 receptor activity / interleukin-8 binding / chemokine receptor activity / negative regulation of cell adhesion molecule production / CXCR chemokine receptor binding / ATF4 activates genes in response to endoplasmic reticulum stress / embryonic digestive tract development ...regulation of single stranded viral RNA replication via double stranded DNA intermediate / regulation of entry of bacterium into host cell / interleukin-8 receptor binding / interleukin-8 receptor activity / interleukin-8 binding / chemokine receptor activity / negative regulation of cell adhesion molecule production / CXCR chemokine receptor binding / ATF4 activates genes in response to endoplasmic reticulum stress / embryonic digestive tract development / neutrophil activation / induction of positive chemotaxis / C-C chemokine receptor activity / C-C chemokine binding / chemokine activity / positive regulation of neutrophil chemotaxis / Chemokine receptors bind chemokines / dendritic cell chemotaxis / negative regulation of G protein-coupled receptor signaling pathway / Interleukin-10 signaling / cellular response to interleukin-1 / regulation of cell adhesion / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / cellular response to fibroblast growth factor stimulus / D2 dopamine receptor binding / response to prostaglandin E / neutrophil chemotaxis / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / secretory granule membrane / Peptide ligand-binding receptors / response to endoplasmic reticulum stress / regulation of mitotic spindle organization / Regulation of insulin secretion / calcium-mediated signaling / response to molecule of bacterial origin / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / receptor internalization / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / chemotaxis / Activation of the phototransduction cascade / positive regulation of angiogenesis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / antimicrobial humoral immune response mediated by antimicrobial peptide / photoreceptor disc membrane / cellular response to tumor necrosis factor / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / heparin binding / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / cellular response to lipopolysaccharide / positive regulation of cytosolic calcium ion concentration / G protein activity / retina development in camera-type eye / GTPase binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
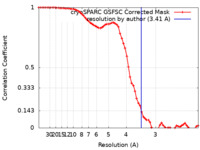
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Ishimoto N / Park JH / Park SY | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of CXC chemokine receptor 1 ligand binding and activation. Authors: Naito Ishimoto / Jae-Hyun Park / Kouki Kawakami / Michiko Tajiri / Kenji Mizutani / Satoko Akashi / Jeremy R H Tame / Asuka Inoue / Sam-Yong Park /  Abstract: Neutrophil granulocytes play key roles in innate immunity and shaping adaptive immune responses. They are attracted by chemokines to sites of infection and tissue damage, where they kill and ...Neutrophil granulocytes play key roles in innate immunity and shaping adaptive immune responses. They are attracted by chemokines to sites of infection and tissue damage, where they kill and phagocytose bacteria. The chemokine CXCL8 (also known as interleukin-8, abbreviated IL-8) and its G-protein-coupled receptors CXCR1 and CXCR2 are crucial elements in this process, and also the development of many cancers. These GPCRs have therefore been the target of many drug development campaigns and structural studies. Here, we solve the structure of CXCR1 complexed with CXCL8 and cognate G-proteins using cryo-EM, showing the detailed interactions between the receptor, the chemokine and Gαi protein. Unlike the closely related CXCR2, CXCR1 strongly prefers to bind CXCL8 in its monomeric form. The model shows that steric clashes would form between dimeric CXCL8 and extracellular loop 2 (ECL2) of CXCR1. Consistently, transplanting ECL2 of CXCR2 onto CXCR1 abolishes the selectivity for the monomeric chemokine. Our model and functional analysis of various CXCR1 mutants will assist efforts in structure-based drug design targeting specific CXC chemokine receptor subtypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35351.map.gz emd_35351.map.gz | 28 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35351-v30.xml emd-35351-v30.xml emd-35351.xml emd-35351.xml | 28.7 KB 28.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35351_fsc.xml emd_35351_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35351.png emd_35351.png | 51.5 KB | ||
| Masks |  emd_35351_msk_1.map emd_35351_msk_1.map | 29.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35351.cif.gz emd-35351.cif.gz | 7.3 KB | ||
| Others |  emd_35351_additional_1.map.gz emd_35351_additional_1.map.gz emd_35351_additional_2.map.gz emd_35351_additional_2.map.gz emd_35351_half_map_1.map.gz emd_35351_half_map_1.map.gz emd_35351_half_map_2.map.gz emd_35351_half_map_2.map.gz | 26.3 MB 14.8 MB 27.5 MB 27.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35351 http://ftp.pdbj.org/pub/emdb/structures/EMD-35351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35351 | HTTPS FTP |
-Related structure data
| Related structure data |  8ic0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35351.map.gz / Format: CCP4 / Size: 29.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35351.map.gz / Format: CCP4 / Size: 29.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07313 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35351_msk_1.map emd_35351_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
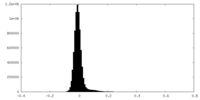
| Density Histograms |
-Additional map: The map was calculated by deepEMhancer
| File | emd_35351_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map was calculated by deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
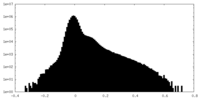
| Density Histograms |
-Additional map: map
| File | emd_35351_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_35351_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_35351_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of human chemokine receptor 1
| Entire | Name: Cryo-EM structure of human chemokine receptor 1 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of human chemokine receptor 1
| Supramolecule | Name: Cryo-EM structure of human chemokine receptor 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: C-X-C chemokine receptor type 1
| Macromolecule | Name: C-X-C chemokine receptor type 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.935199 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNITDPQMW DFDDLNFTGM PPADEDYSPC MLETETLNKY VVIIAYALVF LLSLLGNSLV MLVILYSRVG RSVTDVYLLN LALADLLFA LTLPIWAASK VNGWIFGTFL CKVVSLLKEV NFYSGILLLA CISVDRYLAI VHATRTLTQK RHLVKFVCLG C WGLSMNLS ...String: MSNITDPQMW DFDDLNFTGM PPADEDYSPC MLETETLNKY VVIIAYALVF LLSLLGNSLV MLVILYSRVG RSVTDVYLLN LALADLLFA LTLPIWAASK VNGWIFGTFL CKVVSLLKEV NFYSGILLLA CISVDRYLAI VHATRTLTQK RHLVKFVCLG C WGLSMNLS LPFFLFRQAY HPNNSSPVCY EVLGNDTAKW RMVLRILPHT FGFIVPLFVM LFCYGFTLRT LFKAHMGQKH RA MRVIFAV VLIFLLCWLP YNLVLLADTL MRTQVIQESC ERRNNIGRAL DATEILGFLH SCLNPIIYAF IGQNFRHGFL KIL AMHGLV SKEFLARHRV TSYTSSSVNV SSNLHHHHHH HH UniProtKB: C-X-C chemokine receptor type 1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.728152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.506765 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI LGSAGSAGSA UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.409588 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ |
-Macromolecule #6: Interleukin-8
| Macromolecule | Name: Interleukin-8 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.401807 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SAKELRCQCI KTYSKPFHPK FIKELRVIES GPHCANTEII VKLSDGRELC LDPKENWVQR VVEKFLKRAE NS UniProtKB: Interleukin-8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 50 mM HEPES pH8.0, 100 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP, 0.001% LMNG, 0.0001% CHS, 1 uM IL8 | ||||||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4175 / Average exposure time: 4.7 sec. / Average electron dose: 51.16 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8ic0: |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)