[English] 日本語
 Yorodumi
Yorodumi- EMDB-35345: Cryo-EM structure of the erythromycin-bound motilin receptor-Gq p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / GPCR / erythromycin / motilin receptor / Gq / complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhormone binding / G protein-coupled peptide receptor activity / Peptide ligand-binding receptors / electron transport chain / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway ...hormone binding / G protein-coupled peptide receptor activity / Peptide ligand-binding receptors / electron transport chain / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / periplasmic space / electron transfer activity / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.51 Å | |||||||||
 Authors Authors | You C / Jiang Y / Xu HE / Xu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural basis for motilin and erythromycin recognition by motilin receptor. Authors: Chongzhao You / Yumu Zhang / Youwei Xu / Peiyu Xu / Zhen Li / Huadong Li / Sijie Huang / Zecai Chen / Jingru Li / H Eric Xu / Yi Jiang /  Abstract: Motilin is an endogenous peptide hormone almost exclusively expressed in the human gastrointestinal (GI) tract. It activates the motilin receptor (MTLR), a class A G protein-coupled receptor (GPCR), ...Motilin is an endogenous peptide hormone almost exclusively expressed in the human gastrointestinal (GI) tract. It activates the motilin receptor (MTLR), a class A G protein-coupled receptor (GPCR), and stimulates GI motility. To our knowledge, MTLR is the first GPCR reported to be activated by macrolide antibiotics, such as erythromycin. It has attracted extensive attention as a potential drug target for GI disorders. We report two structures of G-coupled human MTLR bound to motilin and erythromycin. Our structures reveal the recognition mechanism of both ligands and explain the specificity of motilin and ghrelin, a related gut peptide hormone, for their respective receptors. These structures also provide the basis for understanding the different recognition modes of erythromycin by MTLR and ribosome. These findings provide a framework for understanding the physiological regulation of MTLR and guiding drug design targeting MTLR for the treatment of GI motility disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35345.map.gz emd_35345.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35345-v30.xml emd-35345-v30.xml emd-35345.xml emd-35345.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35345.png emd_35345.png | 31.6 KB | ||
| Filedesc metadata |  emd-35345.cif.gz emd-35345.cif.gz | 6.7 KB | ||
| Others |  emd_35345_half_map_1.map.gz emd_35345_half_map_1.map.gz emd_35345_half_map_2.map.gz emd_35345_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35345 http://ftp.pdbj.org/pub/emdb/structures/EMD-35345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35345 | HTTPS FTP |
-Validation report
| Summary document |  emd_35345_validation.pdf.gz emd_35345_validation.pdf.gz | 892.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35345_full_validation.pdf.gz emd_35345_full_validation.pdf.gz | 891.8 KB | Display | |
| Data in XML |  emd_35345_validation.xml.gz emd_35345_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_35345_validation.cif.gz emd_35345_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35345 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35345 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35345 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35345 | HTTPS FTP |
-Related structure data
| Related structure data |  8ibuMC  8ibvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35345.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35345.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35345_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
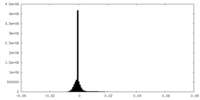
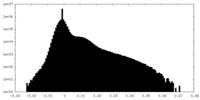
| Density Histograms |
-Half map: #1
| File | emd_35345_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
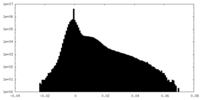
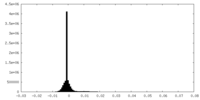
| Density Histograms |
- Sample components
Sample components
-Entire : Erythromycin-bound motilin receptor-Gq protein complex
| Entire | Name: Erythromycin-bound motilin receptor-Gq protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Erythromycin-bound motilin receptor-Gq protein complex
| Supramolecule | Name: Erythromycin-bound motilin receptor-Gq protein complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.724383 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF KSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCSVDT ENARRIFNDC KDIILQMNLR EYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.226992 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWNG SSGGGGSGGG GSSGVSGWRL FKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Soluble cytochrome b562,Motilin receptor,Fusion tag
| Macromolecule | Name: Soluble cytochrome b562,Motilin receptor,Fusion tag / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.667 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLASEN LYFQGGTMGS P WNGSDGPE ...String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLASEN LYFQGGTMGS P WNGSDGPE GAREPPWPAL PPCDERRCSP FPLGALVPVT AVCLCLFVVG VSGNVVTVML IGRYRDMRTT TNLYLGSMAV SD LLILLGL PFDLYRLWRS RPWVFGPLLC RLSLYVGEGC TYATLLHMTA LSVERYLAIC RPLRARVLVT RRRVRALIAV LWA VALLSA GPFLFLVGVE QDPGISVVPG LNGTARIASS PLASSPPLWL SRAPPPSPPS GPETAEAAAL FSRECRPSPA QLGA LRVML WVTTAYFFLP FLCLSILYGL IGRELWSSRR PLRGPAASGR ERGHRQTVRV LLVVVLAFII CWLPFHVGRI IYINT EDSR MMYFSQYFNI VALQLFYLSA SINPILYNLI SKKYRAAAFK LLLARKSRPR GFHRSRDTAG EVAGDTGGDT VGYTET SAN VKTMGGSSGG GGSGGGGSSG VFTLEDFVGD WEQTAAYNLD QVLEQGGVSS LLQNLAVSVT PIQRIVRSGE NALKIDI HV IIPYEGLSAD QMAQIEEVFK VVYPVDDHHF KVILPYGTLV IDGVTPNMLN YFGRPYEGIA VFDGKKITVT GTLWNGNK I IDERLITPDG SMLFRVTINS UniProtKB: Soluble cytochrome b562, Motilin receptor |
-Macromolecule #6: ERYTHROMYCIN A
| Macromolecule | Name: ERYTHROMYCIN A / type: ligand / ID: 6 / Number of copies: 1 / Formula: ERY |
|---|---|
| Molecular weight | Theoretical: 733.927 Da |
| Chemical component information |  ChemComp-ERY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.91 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 8.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.51 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 261108 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)