+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ecCTPS filament bound with CTP, NADH, DON | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / metabolic filament / CTP synthase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoophidium / CTP synthase (glutamine hydrolysing) / CTP synthase activity / 'de novo' CTP biosynthetic process / pyrimidine nucleobase biosynthetic process / ATP binding / metal ion binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Guo CJ / Liu JL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: mLife / Year: 2024 Journal: mLife / Year: 2024Title: Filamentation and inhibition of prokaryotic CTP synthase with ligands. Authors: Chenjun Guo / Zixuan Wang / Ji-Long Liu /   Abstract: Cytidine triphosphate synthase (CTPS) plays a pivotal role in the de novo synthesis of cytidine triphosphate (CTP), a fundamental building block for RNA and DNA that is essential for life. CTPS is ...Cytidine triphosphate synthase (CTPS) plays a pivotal role in the de novo synthesis of cytidine triphosphate (CTP), a fundamental building block for RNA and DNA that is essential for life. CTPS is capable of directly binding to all four nucleotide triphosphates: adenine triphosphate, uridine triphosphate, CTP, and guanidine triphosphate. Furthermore, CTPS can form cytoophidia in vivo and metabolic filaments in vitro, undergoing regulation at multiple levels. CTPS is considered a potential therapeutic target for combating invasions or infections by viral or prokaryotic pathogens. Utilizing cryo-electron microscopy, we determined the structure of CTPS (ecCTPS) filament in complex with CTP, nicotinamide adenine dinucleotide (NADH), and the covalent inhibitor 6-diazo-5-oxo- l-norleucine (DON), achieving a resolution of 2.9 Å. We constructed a phylogenetic tree based on differences in filament-forming interfaces and designed a variant to validate our hypothesis, providing an evolutionary perspective on CTPS filament formation. Our computational analysis revealed a solvent-accessible ammonia tunnel upon DON binding. Through comparative structural analysis, we discern a distinct mode of CTP binding of ecCTPS that differs from eukaryotic counterparts. Combining biochemical assays and structural analysis, we determined and validated the synergistic inhibitory effects of CTP with NADH or adenine on CTPS. Our results expand our comprehension of the diverse regulatory aspects of CTPS and lay a foundation for the design of specific inhibitors targeting prokaryotic CTPS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35278.map.gz emd_35278.map.gz | 44.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35278-v30.xml emd-35278-v30.xml emd-35278.xml emd-35278.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35278_fsc.xml emd_35278_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35278.png emd_35278.png | 51.3 KB | ||
| Masks |  emd_35278_msk_1.map emd_35278_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35278.cif.gz emd-35278.cif.gz | 5.6 KB | ||
| Others |  emd_35278_half_map_1.map.gz emd_35278_half_map_1.map.gz emd_35278_half_map_2.map.gz emd_35278_half_map_2.map.gz | 44.7 MB 44.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35278 http://ftp.pdbj.org/pub/emdb/structures/EMD-35278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35278 | HTTPS FTP |
-Validation report
| Summary document |  emd_35278_validation.pdf.gz emd_35278_validation.pdf.gz | 799.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35278_full_validation.pdf.gz emd_35278_full_validation.pdf.gz | 799 KB | Display | |
| Data in XML |  emd_35278_validation.xml.gz emd_35278_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_35278_validation.cif.gz emd_35278_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35278 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35278 | HTTPS FTP |
-Related structure data
| Related structure data |  8i9oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35278.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35278.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35278_msk_1.map emd_35278_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35278_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35278_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ecCTPS filament with CTP,NADH,DON
| Entire | Name: ecCTPS filament with CTP,NADH,DON |
|---|---|
| Components |
|
-Supramolecule #1: ecCTPS filament with CTP,NADH,DON
| Supramolecule | Name: ecCTPS filament with CTP,NADH,DON / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CTP synthase
| Macromolecule | Name: CTP synthase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.317801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTNYIFVTG GVVSSLGKGI AAASLAAILE ARGLNVTIMK LDPYINVDPG TMSPIQHGEV FVTEDGAETD LDLGHYERFI RTKMSRRNN FTTGRIYSDV LRKERRGDYL GATVQVIPHI TNAIKERVLE GGEGHDVVLV EIGGTVGDIE SLPFLEAIRQ M AVEIGREH ...String: MTTNYIFVTG GVVSSLGKGI AAASLAAILE ARGLNVTIMK LDPYINVDPG TMSPIQHGEV FVTEDGAETD LDLGHYERFI RTKMSRRNN FTTGRIYSDV LRKERRGDYL GATVQVIPHI TNAIKERVLE GGEGHDVVLV EIGGTVGDIE SLPFLEAIRQ M AVEIGREH TLFMHLTLVP YMAASGEVKT KPTQHSVKEL LSIGIQPDIL ICRSDRAVPA NERAKIALFC NVPEKAVISL KD VDSIYKI PGLLKSQGLD DYICKRFSLN CPEANLSEWE QVIFEEANPV SEVTIGMVGK YIELPDAYKS VIEALKHGGL KNR VSVNIK LIDSQDVETR GVEILKGLDA ILVPGGFGYR GVEGMITTAR FARENNIPYL GICLGMQVAL IDYARHVANM ENAN STEFV PDCKYPVVAL ITEWRDENGN VEVRSEKSDL GGTMRLGAQQ CQLVDDSLVR QLYNAPTIVE RHRHRYEVNN MLLKQ IEDA GLRVAGRSGD DQLVEIIEVP NHPWFVACQF HPEFTSTPRD GHPLFAGFVK AASEFQKRQA UniProtKB: CTP synthase |
-Macromolecule #2: ADENINE
| Macromolecule | Name: ADENINE / type: ligand / ID: 2 / Number of copies: 4 / Formula: ADE |
|---|---|
| Molecular weight | Theoretical: 135.127 Da |
| Chemical component information |  ChemComp-ADE: |
-Macromolecule #3: 5-OXO-L-NORLEUCINE
| Macromolecule | Name: 5-OXO-L-NORLEUCINE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ONL |
|---|---|
| Molecular weight | Theoretical: 145.156 Da |
| Chemical component information | 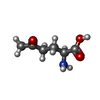 ChemComp-ONL: |
-Macromolecule #4: CYTIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: CYTIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: CTP |
|---|---|
| Molecular weight | Theoretical: 483.156 Da |
| Chemical component information |  ChemComp-CTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































