[English] 日本語
 Yorodumi
Yorodumi- EMDB-35251: Density map of the OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulf... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density map of the OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulfide-bonded to MlaC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteria / outer membrane / phospholipid / lipid asymmetry / membrane protein / protein complex structure / channel / LIPID TRANSPORT | |||||||||
| Biological species |  | |||||||||
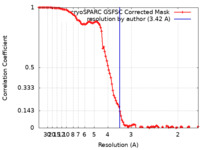
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Yeow J / Luo M / Chng SS | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular mechanism of phospholipid transport at the bacterial outer membrane interface. Authors: Jiang Yeow / Min Luo / Shu-Sin Chng /  Abstract: The outer membrane (OM) of Gram-negative bacteria is an asymmetric lipid bilayer with outer leaflet lipopolysaccharides and inner leaflet phospholipids (PLs). This unique lipid asymmetry renders the ...The outer membrane (OM) of Gram-negative bacteria is an asymmetric lipid bilayer with outer leaflet lipopolysaccharides and inner leaflet phospholipids (PLs). This unique lipid asymmetry renders the OM impermeable to external insults, including antibiotics and bile salts. To maintain this barrier, the OmpC-Mla system removes mislocalized PLs from the OM outer leaflet, and transports them to the inner membrane (IM); in the first step, the OmpC-MlaA complex transfers PLs to the periplasmic chaperone MlaC, but mechanistic details are lacking. Here, we biochemically and structurally characterize the MlaA-MlaC transient complex. We map the interaction surfaces between MlaA and MlaC in Escherichia coli, and show that electrostatic interactions are important for MlaC recruitment to the OM. We further demonstrate that interactions with MlaC modulate conformational states in MlaA. Finally, we solve a 2.9-Å cryo-EM structure of a disulfide-trapped OmpC-MlaA-MlaC complex in nanodiscs, reinforcing the mechanism of MlaC recruitment, and highlighting membrane thinning as a plausible strategy for directing lipids for transport. Our work offers critical insights into retrograde PL transport by the OmpC-Mla system in maintaining OM lipid asymmetry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35251.map.gz emd_35251.map.gz | 62.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35251-v30.xml emd-35251-v30.xml emd-35251.xml emd-35251.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35251_fsc.xml emd_35251_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_35251.png emd_35251.png | 107.1 KB | ||
| Masks |  emd_35251_msk_1.map emd_35251_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35251.cif.gz emd-35251.cif.gz | 4.9 KB | ||
| Others |  emd_35251_half_map_1.map.gz emd_35251_half_map_1.map.gz emd_35251_half_map_2.map.gz emd_35251_half_map_2.map.gz | 116 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35251 http://ftp.pdbj.org/pub/emdb/structures/EMD-35251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35251 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35251.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35251.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35251_msk_1.map emd_35251_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35251_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35251_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulfide-bonded to MlaC
| Entire | Name: OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulfide-bonded to MlaC |
|---|---|
| Components |
|
-Supramolecule #1: OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulfide-bonded to MlaC
| Supramolecule | Name: OmpC3-MlaA3 complex in MSP2N2 nanodiscs disulfide-bonded to MlaC type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Outer membrane complex of 3OmpC with disulfide-trapped 3MlaA and 3MlaC reconstituted in Escherichia coli polar lipids and MSP2N2 nanodiscs |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28 KDa |
-Supramolecule #2: Outer membrane porin C
| Supramolecule | Name: Outer membrane porin C / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Trimeric OmpC porins in complex with MlaA at the outer membrane disulfide-trapped with MlaC |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Complex of MlaA disulfide trapped with MlaC
| Supramolecule | Name: Complex of MlaA disulfide trapped with MlaC / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 Details: Intermembrane phospholipid transport system lipoprotein MlaA disulfide trapped with MlaC |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Tris-buffered saline (TBS) buffer (20 mM Tris HCl pH 8.0, 150 mM NaCl) | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Tridiem 4K / Energy filter - Slit width: 20 eV Details: Gatan GIF post-column energy filter operated in zero-loss mode |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6018 / Average exposure time: 5.99 sec. / Average electron dose: 90.0 e/Å2 Details: Images were collected in movie-mode at 50 frames per image |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Overall B value: 50.2 |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)