[English] 日本語
 Yorodumi
Yorodumi- EMDB-35242: The helical cylinder of Autographa californica multiple nucleopol... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The helical cylinder of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) nucleocapsid | |||||||||
 Map data Map data | cryo-em structure of AcMNPV helical body | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / helical / VIRAL PROTEIN | |||||||||
| Biological species |  Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus | |||||||||
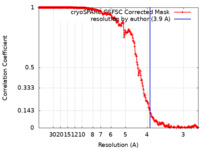
| Method | helical reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Jia X / Gao Y / Zhang Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Architecture of the baculovirus nucleocapsid revealed by cryo-EM. Authors: Xudong Jia / Yuanzhu Gao / Yuxuan Huang / Linjun Sun / Siduo Li / Hongmei Li / Xueqing Zhang / Yinyin Li / Jian He / Wenbi Wu / Harikanth Venkannagari / Kai Yang / Matthew L Baker / Qinfen Zhang /   Abstract: Baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been widely used as a bioinsecticide and a protein expression vector. Despite their importance, very little is known ...Baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been widely used as a bioinsecticide and a protein expression vector. Despite their importance, very little is known about the structure of most baculovirus proteins. Here, we show a 3.2 Å resolution structure of helical cylindrical body of the AcMNPV nucleocapsid, composed of VP39, as well as 4.3 Å resolution structures of both the head and the base of the nucleocapsid composed of over 100 protein subunits. AcMNPV VP39 demonstrates some features of the HK97-like fold and utilizes disulfide-bonds and a set of interactions at its C-termini to mediate nucleocapsid assembly and stability. At both ends of the nucleocapsid, the VP39 cylinder is constricted by an outer shell ring composed of proteins AC104, AC142 and AC109. AC101(BV/ODV-C42) and AC144(ODV-EC27) form a C14 symmetric inner layer at both capsid head and base. In the base, these proteins interact with a 7-fold symmetric capsid plug, while a portal-like structure is seen in the central portion of head. Additionally, we propose an application of AlphaFold2 for model building in intermediate resolution density. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35242.map.gz emd_35242.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35242-v30.xml emd-35242-v30.xml emd-35242.xml emd-35242.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35242_fsc.xml emd_35242_fsc.xml | 19.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35242.png emd_35242.png | 290 KB | ||
| Masks |  emd_35242_msk_1.map emd_35242_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35242.cif.gz emd-35242.cif.gz | 4.8 KB | ||
| Others |  emd_35242_half_map_1.map.gz emd_35242_half_map_1.map.gz emd_35242_half_map_2.map.gz emd_35242_half_map_2.map.gz | 752.8 MB 752.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35242 http://ftp.pdbj.org/pub/emdb/structures/EMD-35242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35242 | HTTPS FTP |
-Validation report
| Summary document |  emd_35242_validation.pdf.gz emd_35242_validation.pdf.gz | 956.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35242_full_validation.pdf.gz emd_35242_full_validation.pdf.gz | 956.2 KB | Display | |
| Data in XML |  emd_35242_validation.xml.gz emd_35242_validation.xml.gz | 29.2 KB | Display | |
| Data in CIF |  emd_35242_validation.cif.gz emd_35242_validation.cif.gz | 38.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35242 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35242 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35242.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35242.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-em structure of AcMNPV helical body | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35242_msk_1.map emd_35242_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_35242_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_35242_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Autographa californica multiple nucleopolyhedrovirus
| Entire | Name:  Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
|---|---|
| Components |
|
-Supramolecule #1: Autographa californica multiple nucleopolyhedrovirus
| Supramolecule | Name: Autographa californica multiple nucleopolyhedrovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 307456 Sci species name: Autographa californica multiple nucleopolyhedrovirus Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: major viral capsid protein
| Macromolecule | Name: major viral capsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Sequence | String: MALVPVGMAP RQMRVNRCIF ASIVSFDACI TYKSPCSPDA YHDD GWFIC NNHLIKRFKM SKMVLPIFDE DDNQFKMTIA RHLVGNKERG IKRILIPSAT NYQ DVFNLN SMMQAEQLIF HLIYNNENAV NTICDNLKYT EGFTSNTQRV IHSVYATTKS IL DTTNPNT ...String: MALVPVGMAP RQMRVNRCIF ASIVSFDACI TYKSPCSPDA YHDD GWFIC NNHLIKRFKM SKMVLPIFDE DDNQFKMTIA RHLVGNKERG IKRILIPSAT NYQ DVFNLN SMMQAEQLIF HLIYNNENAV NTICDNLKYT EGFTSNTQRV IHSVYATTKS IL DTTNPNT FCSRVSRDEL RFFDVTNARA LRGGAGDQLF NNYSGFLQNL IRRAVAPEYL Q IDTEELRF RNCATCIIDE TGLVASVPDG PELYNPIRSS DIMRSQPNRL QIRNVLKFEG DTRELDRTL SGYEEYPTYV PLFLGYQIIN SENNFLRNDF IPRANPNATL GGGAVAGPA PGVAGEAGGG IAV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)