[English] 日本語
 Yorodumi
Yorodumi- EMDB-35208: Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | polyphosphate polymerase / VTC complex / coupled synthesis and translocation / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvacuolar transporter chaperone complex / engulfment of target by autophagosome / ATP-polyphosphate phosphotransferase / polyphosphate biosynthetic process / polyphosphate kinase activity / microautophagy / vacuole fusion, non-autophagic / polyphosphate metabolic process / vacuolar transport / intracellular phosphate ion homeostasis ...vacuolar transporter chaperone complex / engulfment of target by autophagosome / ATP-polyphosphate phosphotransferase / polyphosphate biosynthetic process / polyphosphate kinase activity / microautophagy / vacuole fusion, non-autophagic / polyphosphate metabolic process / vacuolar transport / intracellular phosphate ion homeostasis / fungal-type vacuole membrane / inositol hexakisphosphate binding / vacuolar membrane / autophagosome membrane / cell periphery / cytoplasmic vesicle / cell cortex / nuclear membrane / calmodulin binding / mRNA binding / endoplasmic reticulum membrane / endoplasmic reticulum Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
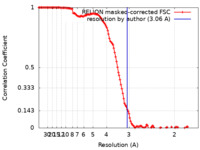
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||||||||
 Authors Authors | Mayer A / Wu S / Ye S | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit. Authors: Wei Liu / Jiening Wang / Véronique Comte-Miserez / Mengyu Zhang / Xuejing Yu / Qingfeng Chen / Henning Jacob Jessen / Andreas Mayer / Shan Wu / Sheng Ye /    Abstract: The eukaryotic vacuolar transporter chaperone (VTC) complex acts as a polyphosphate (polyP) polymerase that synthesizes polyP from adenosine triphosphate (ATP) and translocates polyP across the ...The eukaryotic vacuolar transporter chaperone (VTC) complex acts as a polyphosphate (polyP) polymerase that synthesizes polyP from adenosine triphosphate (ATP) and translocates polyP across the vacuolar membrane to maintain an intracellular phosphate (P ) homeostasis. To discover how the VTC complex performs its function, we determined a cryo-electron microscopy structure of an endogenous VTC complex (Vtc4/Vtc3/Vtc1) purified from Saccharomyces cerevisiae at 3.1 Å resolution. The structure reveals a heteropentameric architecture of one Vtc4, one Vtc3, and three Vtc1 subunits. The transmembrane region forms a polyP-selective channel, likely adopting a resting state conformation, in which a latch-like, horizontal helix of Vtc4 limits the entrance. The catalytic Vtc4 central domain is located on top of the pseudo-symmetric polyP channel, creating a strongly electropositive pathway for nascent polyP that can couple synthesis to translocation. The SPX domain of the catalytic Vtc4 subunit positively regulates polyP synthesis by the VTC complex. The noncatalytic Vtc3 regulates VTC through a phosphorylatable loop. Our findings, along with the functional data, allow us to propose a mechanism of polyP channel gating and VTC complex activation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35208.map.gz emd_35208.map.gz | 166.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35208-v30.xml emd-35208-v30.xml emd-35208.xml emd-35208.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35208_fsc.xml emd_35208_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35208.png emd_35208.png | 156.3 KB | ||
| Filedesc metadata |  emd-35208.cif.gz emd-35208.cif.gz | 7.9 KB | ||
| Others |  emd_35208_additional_1.map.gz emd_35208_additional_1.map.gz emd_35208_half_map_1.map.gz emd_35208_half_map_1.map.gz emd_35208_half_map_2.map.gz emd_35208_half_map_2.map.gz | 155.9 MB 140.4 MB 140.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35208 http://ftp.pdbj.org/pub/emdb/structures/EMD-35208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35208 | HTTPS FTP |
-Related structure data
| Related structure data |  8i6vMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35208.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35208.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.851 Å | ||||||||||||||||||||||||||||||||||||
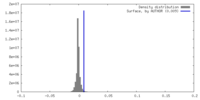
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_35208_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
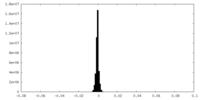
| Density Histograms |
-Half map: #2
| File | emd_35208_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
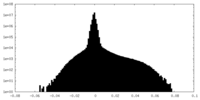
| Density Histograms |
-Half map: #1
| File | emd_35208_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
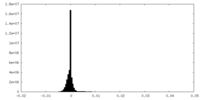
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc...
| Entire | Name: Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc...
| Supramolecule | Name: Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vacuolar transporter chaperone complex subunit 1
| Macromolecule | Name: Vacuolar transporter chaperone complex subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.388053 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSAPLLQRT PGKKIALPTR VEPKVFFANE RTFLSWLNFT VMLGGLGVGL LNFGDKIGRV SAGLFTFVAM GTMIYALVTY HWRAAAIRR RGSGPYDDRL GPTLLCFFLL VAVIINFILR LKYNDANTKL UniProtKB: Vacuolar transporter chaperone complex subunit 1 |
-Macromolecule #2: Vacuolar transporter chaperone 3 complex subunit 3
| Macromolecule | Name: Vacuolar transporter chaperone 3 complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.684312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLFGIKLAND VYPPWKDSYI DYERLKKLLK ESVIHDGRSS VDSWSERNES DFVEALDKEL EKVYTFQISK YNAVLRKLDD LEENTKSAE KIQKINSEQF KNTLEECLDE AQRLDNFDRL NFTGFIKIVK KHDKLHPNYP SVKSLLQVRL KELPFNNSEE Y SPLLYRIS ...String: MLFGIKLAND VYPPWKDSYI DYERLKKLLK ESVIHDGRSS VDSWSERNES DFVEALDKEL EKVYTFQISK YNAVLRKLDD LEENTKSAE KIQKINSEQF KNTLEECLDE AQRLDNFDRL NFTGFIKIVK KHDKLHPNYP SVKSLLQVRL KELPFNNSEE Y SPLLYRIS YLYEFLRSNY DHPNTVSKSL ASTSKLSHFS NLEDASFKSY KFWVHDDNIM EVKARILRHL PALVYASVPN EN DDFVDNL ESDVRVQPEA RLNIGSKSNS LSSDGNSNQD VEIGKSKSVI FPQSYDPTIT TLYFDNDFFD LYNNRLLKIS GAP TLRLRW IGKLLDKPDI FLEKRTFTEN TETGNSSFEE IRLQMKAKFI NNFIFKNDPS YKNYLINQLR ERGTQKEELE KLSR DFDNI QNFIVEEKLQ PVLRATYNRT AFQIPGDQSI RVTIDSNIMY IREDSLDKNR PIRNPENWHR DDIDSNIPNP LRFLR AGEY SKFPYSVMEI KVINQDNSQM PNYEWIKDLT NSHLVNEVPK FSLYLQGVAS LFGEDDKYVN ILPFWLPDLE TDIRKN PQE AYEEEKKTLQ KQKSIHDKLD NMRRLSKISV PDGKTTERQG QKDQNTRHVI ADLEDHESSD EEGTALPKKS AVKKGKK FK TNAAFLKILA GKNISENGND PYSDDTDSAS SFQLPPGVKK PVHLLKNAGP VKVEAKVWLA NERTFNRWLS VTTLLSVL T FSIYNSVQKA EFPQLADLLA YVYFFLTLFC GVWAYRTYLK RLTLIKGRSG KHLDAPVGPI LVAVVLIVTL VVNFSVAFK EAARRERGLV NVSSQPSLPR TLKPIQDFIF NLVGE UniProtKB: Vacuolar transporter chaperone 3 complex subunit 3 |
-Macromolecule #3: Vacuolar transporter chaperone complex subunit 4
| Macromolecule | Name: Vacuolar transporter chaperone complex subunit 4 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: ATP-polyphosphate phosphotransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 83.268664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKFGEHLSKS LIRQYSYYYI SYDDLKTELE DNLSKNNGQW TQELETDFLE SLEIELDKVY TFCKVKHSEV FRRVKEVQEQ VQHTVRLLD SNNPPTQLDF EILEEELSDI IADVHDLAKF SRLNYTGFQK IIKKHDKKTG FILKPVFQVR LDSKPFFKEN Y DELVVKIS ...String: MKFGEHLSKS LIRQYSYYYI SYDDLKTELE DNLSKNNGQW TQELETDFLE SLEIELDKVY TFCKVKHSEV FRRVKEVQEQ VQHTVRLLD SNNPPTQLDF EILEEELSDI IADVHDLAKF SRLNYTGFQK IIKKHDKKTG FILKPVFQVR LDSKPFFKEN Y DELVVKIS QLYDIARTSG RPIKGDSSAG GKQQNFVRQT TKYWVHPDNI TELKLIILKH LPVLVFNTNK EFEREDSAIT SI YFDNENL DLYYGRLRKD EGAEAHRLRW YGGMSTDTIF VERKTHREDW TGEKSVKARF ALKERHVNDF LKGKYTVDQV FAK MRKEGK KPMNEIENLE ALASEIQYVM LKKKLRPVVR SFYNRTAFQL PGDARVRISL DTELTMVRED NFDGVDRTHK NWRR TDIGV DWPFKQLDDK DICRFPYAVL EVKLQTQLGQ EPPEWVRELV GSHLVEPVPK FSKFIHGVAT LLNDKVDSIP FWLPQ MDVD IRKPPLPTNI EITRPGRSDN EDNDFDEDDE DDAALVAAMT NAPGNSLDIE ESVGYGATSA PTSNTNHVVE SANAAY YQR KIRNAENPIS KKYYEIVAFF DHYFNGDQIS KIPKGTTFDT QIRAPPGKTI CVPVRVEPKV YFATERTYLS WLSISIL LG GVSTTLLTYG SPTAMIGSIG FFITSLAVLI RTVMVYAKRV VNIRLKRAVD YEDKIGPGMV SVFLILSILF SFFCNLVA K UniProtKB: Vacuolar transporter chaperone complex subunit 4 |
-Macromolecule #4: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 4 / Number of copies: 1 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #5: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #6: TRIPHOSPHATE
| Macromolecule | Name: TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: 3PO |
|---|---|
| Molecular weight | Theoretical: 257.955 Da |
| Chemical component information |  ChemComp-3PO: |
-Macromolecule #7: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)