+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CalA3 complex structure with amidation product | |||||||||
 Map data Map data | postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | megaenzyme / HYDROLASE / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolyketide biosynthetic process / beta-ketoacyl-[acyl-carrier-protein] synthase I / fatty acid synthase activity / 3-oxoacyl-[acyl-carrier-protein] synthase activity / antibiotic biosynthetic process / fatty acid biosynthetic process Similarity search - Function | |||||||||
| Biological species |  Streptomyces chartreusis NRRL 3882cha (bacteria) / Streptomyces chartreusis NRRL 3882cha (bacteria) /  Streptomyces chartreusis NRRL 3882 (bacteria) Streptomyces chartreusis NRRL 3882 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | |||||||||
 Authors Authors | Wang J / Wang Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: C-N bond formation by a polyketide synthase. Authors: Jialiang Wang / Xiaojie Wang / Xixi Li / LiangLiang Kong / Zeqian Du / Dandan Li / Lixia Gou / Hao Wu / Wei Cao / Xiaozheng Wang / Shuangjun Lin / Ting Shi / Zixin Deng / Zhijun Wang / Jingdan Liang /  Abstract: Assembly-line polyketide synthases (PKSs) are molecular factories that produce diverse metabolites with wide-ranging biological activities. PKSs usually work by constructing and modifying the ...Assembly-line polyketide synthases (PKSs) are molecular factories that produce diverse metabolites with wide-ranging biological activities. PKSs usually work by constructing and modifying the polyketide backbone successively. Here, we present the cryo-EM structure of CalA3, a chain release PKS module without an ACP domain, and its structures with amidation or hydrolysis products. The domain organization reveals a unique "∞"-shaped dimeric architecture with five connected domains. The catalytic region tightly contacts the structural region, resulting in two stabilized chambers with nearly perfect symmetry while the N-terminal docking domain is flexible. The structures of the ketosynthase (KS) domain illustrate how the conserved key residues that canonically catalyze C-C bond formation can be tweaked to mediate C-N bond formation, revealing the engineering adaptability of assembly-line polyketide synthases for the production of novel pharmaceutical agents. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35188.map.gz emd_35188.map.gz | 13.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35188-v30.xml emd-35188-v30.xml emd-35188.xml emd-35188.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35188_fsc.xml emd_35188_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35188.png emd_35188.png | 108.8 KB | ||
| Filedesc metadata |  emd-35188.cif.gz emd-35188.cif.gz | 7.3 KB | ||
| Others |  emd_35188_additional_1.map.gz emd_35188_additional_1.map.gz emd_35188_half_map_1.map.gz emd_35188_half_map_1.map.gz emd_35188_half_map_2.map.gz emd_35188_half_map_2.map.gz | 167 MB 170 MB 170 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35188 http://ftp.pdbj.org/pub/emdb/structures/EMD-35188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35188 | HTTPS FTP |
-Related structure data
| Related structure data |  8i4yMC  7wvzC  8i4zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35188.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35188.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: full map
| File | emd_35188_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_35188_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_35188_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : an assembly-line polyketide synthase module containing KS-AT-DH-K...
| Entire | Name: an assembly-line polyketide synthase module containing KS-AT-DH-KR domains with amidation product |
|---|---|
| Components |
|
-Supramolecule #1: an assembly-line polyketide synthase module containing KS-AT-DH-K...
| Supramolecule | Name: an assembly-line polyketide synthase module containing KS-AT-DH-KR domains with amidation product type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Streptomyces chartreusis NRRL 3882cha (bacteria) Streptomyces chartreusis NRRL 3882cha (bacteria) |
| Molecular weight | Theoretical: 0.36 kDa/nm |
-Macromolecule #1: Beta-ketoacyl-acyl-carrier-protein synthase I
| Macromolecule | Name: Beta-ketoacyl-acyl-carrier-protein synthase I / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces chartreusis NRRL 3882 (bacteria) Streptomyces chartreusis NRRL 3882 (bacteria) |
| Molecular weight | Theoretical: 178.383562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPDEEKLQKY LRKVTAELQQ ARRRLAAAES QSQEPIAVLG IGCRFPGGVR SPEDLWDLVD SGGDAVGGLP AGRGWQAGSA LDGVNAGFI HGVEEFDPYF FGLDPVEAAA MDPQQRLLLE TTWEAFERAG IDPVAARGSR TAVYAGVQFG GYPLLMREAP P PQVLDHLG ...String: MPDEEKLQKY LRKVTAELQQ ARRRLAAAES QSQEPIAVLG IGCRFPGGVR SPEDLWDLVD SGGDAVGGLP AGRGWQAGSA LDGVNAGFI HGVEEFDPYF FGLDPVEAAA MDPQQRLLLE TTWEAFERAG IDPVAARGSR TAVYAGVQFG GYPLLMREAP P PQVLDHLG LGNSVGAASG RLAYQFGLLG GAVTVDTQCT SSIVALHLAV KALRNGECAL ALAGGACVMS LPTVLMDFHR RS LLAPDGR SKSFAAAADG VSLAEGAGML LLERLSDARR NGHPVMAVIR GTAINQDGAT NGIISPSGRA QERVIRAALA DGR VTADSV DAVEGHGVGA TLGDGVEVTS LLSTYGQERP AGRPLLLGSV KSNIGHTQTV GAVAGIVKLV MALRNERLPR TVHV DGPTP HADWSSGTVR LLTEPEPWRR GERVRRAGLT CLTLSGTNGH LILEEPPADE PAARPANPER TVPLVLSAKS PTALR EQAE RLRATITAAE PVDVGHSLHT TRSSFRHRAV VLGTGREELA AGLDALAGDR TADGLVRGVA RAQGQTALLF GGAGDG TSG DRPADAEGPR TARGLYEAFP AFAEALDEVT EHLAGLLGPE VRAAVREPGP ACAEPTVVGQ AVAFALNTAL HRLLTAF AV RPDATLGHGA GEVAAAYAAG ALSLADGAAL VTALGRITER VATGPGASVW VRATEDEVRA ALSGSQEQVG AAVAAVDE P GTTVVSGDAG AVARVAAHWR AHGRATGAPR PARLLLSPDD EQAALAELRA IVAGLAFREP EVPLLSTVTG QPVEPAELR SAEHWLDHLR GPTRFLDGVR RLRTDGVTRL VGLDLSGDLT GPAGRSAAGF GEPGRPLLLA SVPGGGRPPG QALLSALGEL HTDGVAIDW SQAFEGRGAR RVDLPTYPYQ KVRCWLVPPE PQVSVVAAPP HPLLGTALDL VDATGQSFTQ QLTPGQVAGV F GQQLYGTP VLPAGARLEW LLAAARHGSP DSAWTLTGIR LPGTVSAASG TPVALQTSRE DSGDGHRVRA FVKGPGTGGG RW AERGGAT VVPAVTRPAP DRVDPESLPE GLAELDVAEV YRRLWRQGSD YAEPLRVLRR VWLGGDEAVA LVGTADVPTG PSG WSRWAA VLEAAVQLAA LSGSGPRTPV SVDRLEVSGP PSEVVWLRVR HGADGAADAV VLSGEGVRLA AVQGLRLRPM AGRE PAGLA EAPLERHEVV WHALAEDGRP GAIGGGTGSW LVFSDDPERA AAWCDELALF GVPAVALAGE DAEGRDGTET VPVGT GDPD VVGKTFAELR ERGVTVAGLL VHDAGDAREP ASGADDPLDA ACRRGGRTLA LVRGFLQEYA EQTPRIVLCS AGAAAG LAG GPPHPAQAPL TALFTSLVWE HPELPCAQVD LDPAEDPPTV VSLLGQVMRL PGAGRLAVRG GRWFEARLER RPAPADR GE RLALRPDATY LVAGGDTRHA AAALEWLAAR GARSVVLAGA ESERGDLAGA RTTGHAGIER LEHVAVDLSS AADVARLA E LCADGRPPLR GVLLLPQPVA GGGLDELDGA RFGAELAGAL RGPVELTRRF TDVGLTGGTD FFVLSTSVVS LPGRAGTVV GSAADAFLTA LARHHRQAGL PVVAAAWGPW LESVDESDEA PAVAFAEAGV YPAPGGEMLD ALLPLPAAGE ADGSGEAGLA RVDWDRYLT AGHRPLPYTV LETRASYDEE KAPGFGQNRM UniProtKB: Beta-ketoacyl-acyl-carrier-protein synthase I |
-Macromolecule #2: 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid
| Macromolecule | Name: 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: ONF |
|---|---|
| Molecular weight | Theoretical: 265.348 Da |
| Chemical component information | 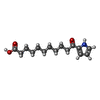 ChemComp-ONF: |
-Macromolecule #3: 3-HYDROXYANTHRANILIC ACID
| Macromolecule | Name: 3-HYDROXYANTHRANILIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: 3HA |
|---|---|
| Molecular weight | Theoretical: 153.135 Da |
| Chemical component information |  ChemComp-3HA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.06 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































