+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of human post-Bact complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spliceosome / Bact complex / RNA splicing / PRP2 / branching / SPLICING | |||||||||
| Function / homology |  Function and homology information Function and homology informationU11/U12 snRNP / regulation of retinoic acid receptor signaling pathway / post-mRNA release spliceosomal complex / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / 3'-5' RNA helicase activity / U7 snRNP / generation of catalytic spliceosome for first transesterification step / B-WICH complex ...U11/U12 snRNP / regulation of retinoic acid receptor signaling pathway / post-mRNA release spliceosomal complex / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / 3'-5' RNA helicase activity / U7 snRNP / generation of catalytic spliceosome for first transesterification step / B-WICH complex / regulation of vitamin D receptor signaling pathway / histone pre-mRNA 3'end processing complex / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / protein methylation / splicing factor binding / U12-type spliceosomal complex / embryonic brain development / methylosome / nuclear retinoic acid receptor binding / 7-methylguanosine cap hypermethylation / positive regulation of androgen receptor activity / Prp19 complex / poly(A) binding / U1 snRNP binding / pICln-Sm protein complex / mRNA 3'-end processing / sno(s)RNA-containing ribonucleoprotein complex / snRNP binding / U2-type catalytic step 1 spliceosome / RNA splicing, via transesterification reactions / small nuclear ribonucleoprotein complex / pre-mRNA binding / regulation of mRNA splicing, via spliceosome / C2H2 zinc finger domain binding / SMN-Sm protein complex / telomerase RNA binding / spliceosomal tri-snRNP complex / telomerase holoenzyme complex / U2-type spliceosomal complex / U2-type precatalytic spliceosome / positive regulation by host of viral transcription / mRNA cis splicing, via spliceosome / P granule / positive regulation of vitamin D receptor signaling pathway / commitment complex / U2-type prespliceosome assembly / nuclear vitamin D receptor binding / Transport of Mature mRNA derived from an Intron-Containing Transcript / U2-type catalytic step 2 spliceosome / Notch binding / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / U4 snRNP / positive regulation of mRNA splicing, via spliceosome / RUNX3 regulates NOTCH signaling / U2 snRNP / SAGA complex / NOTCH4 Intracellular Domain Regulates Transcription / RNA Polymerase II Transcription Termination / U1 snRNP / positive regulation of transcription by RNA polymerase III / pattern recognition receptor activity / NOTCH3 Intracellular Domain Regulates Transcription / ubiquitin-ubiquitin ligase activity / positive regulation of neurogenesis / WD40-repeat domain binding / U2-type prespliceosome / lipid biosynthetic process / nuclear androgen receptor binding / precatalytic spliceosome / K63-linked polyubiquitin modification-dependent protein binding / cyclosporin A binding / positive regulation of transcription by RNA polymerase I / Notch-HLH transcription pathway / positive regulation of transforming growth factor beta receptor signaling pathway / Formation of paraxial mesoderm / mRNA Splicing - Minor Pathway / spliceosomal complex assembly / SMAD binding / mitotic G2 DNA damage checkpoint signaling / regulation of RNA splicing / protein K63-linked ubiquitination / protein peptidyl-prolyl isomerization / blastocyst development / protein localization to nucleus / spliceosomal tri-snRNP complex assembly / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / U5 snRNA binding / U5 snRNP / transcription-coupled nucleotide-excision repair / positive regulation of G1/S transition of mitotic cell cycle / embryonic organ development / proteasomal protein catabolic process / positive regulation of viral genome replication / retinoic acid receptor signaling pathway / U2 snRNA binding / U6 snRNA binding / spliceosomal snRNP assembly / regulation of DNA repair / Cajal body Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhan X / Lu Y / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular basis for the activation of human spliceosome. Authors: Xiechao Zhan / Yichen Lu / Yigong Shi /  Abstract: The spliceosome executes pre-mRNA splicing through four sequential stages: assembly, activation, catalysis, and disassembly. Activation of the spliceosome, namely remodeling of the pre-catalytic ...The spliceosome executes pre-mRNA splicing through four sequential stages: assembly, activation, catalysis, and disassembly. Activation of the spliceosome, namely remodeling of the pre-catalytic spliceosome (B complex) into the activated spliceosome (B complex) and the catalytically activated spliceosome (B complex), involves major flux of protein components and structural rearrangements. Relying on a splicing inhibitor, we have captured six intermediate states between the B and B complexes: pre-B, B-I, B-II, B-III, B-IV, and post-B. Their cryo-EM structures, together with an improved structure of the catalytic step I spliceosome (C complex), reveal how the catalytic center matures around the internal stem loop of U6 snRNA, how the branch site approaches 5'-splice site, how the RNA helicase PRP2 rearranges to bind pre-mRNA, and how U2 snRNP undergoes remarkable movement to facilitate activation. We identify a previously unrecognized key role of PRP2 in spliceosome activation. Our study recapitulates a molecular choreography of the human spliceosome during its catalytic activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35111.map.gz emd_35111.map.gz | 398.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35111-v30.xml emd-35111-v30.xml emd-35111.xml emd-35111.xml | 69.6 KB 69.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35111.png emd_35111.png | 76.4 KB | ||
| Filedesc metadata |  emd-35111.cif.gz emd-35111.cif.gz | 21.3 KB | ||
| Others |  emd_35111_half_map_1.map.gz emd_35111_half_map_1.map.gz emd_35111_half_map_2.map.gz emd_35111_half_map_2.map.gz | 391 MB 391 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35111 http://ftp.pdbj.org/pub/emdb/structures/EMD-35111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35111 | HTTPS FTP |
-Validation report
| Summary document |  emd_35111_validation.pdf.gz emd_35111_validation.pdf.gz | 922.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35111_full_validation.pdf.gz emd_35111_full_validation.pdf.gz | 922.2 KB | Display | |
| Data in XML |  emd_35111_validation.xml.gz emd_35111_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_35111_validation.cif.gz emd_35111_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35111 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35111 | HTTPS FTP |
-Related structure data
| Related structure data |  8i0vMC  8i0pC  8i0rC  8i0sC  8i0tC  8i0uC  8i0wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35111.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35111.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.077 Å | ||||||||||||||||||||||||||||||||||||
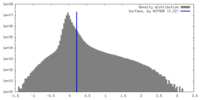
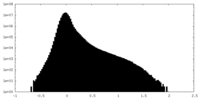
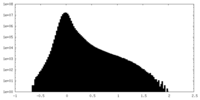
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
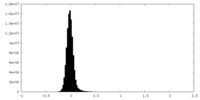
| Density Histograms |
-Half map: #1
| File | emd_35111_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
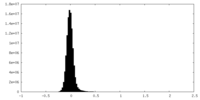
| Density Histograms |
- Sample components
Sample components
+Entire : The human Bact-III complex
+Supramolecule #1: The human Bact-III complex
+Macromolecule #1: Pre-mRNA-processing-splicing factor 8
+Macromolecule #3: 116 kDa U5 small nuclear ribonucleoprotein component
+Macromolecule #4: U5 small nuclear ribonucleoprotein 40 kDa protein
+Macromolecule #8: Pre-mRNA-splicing factor SYF1
+Macromolecule #9: Crooked neck-like protein 1
+Macromolecule #10: Cell division cycle 5-like protein
+Macromolecule #11: Pre-mRNA-splicing factor SYF2
+Macromolecule #12: Protein BUD31 homolog
+Macromolecule #13: Pre-mRNA-splicing factor RBM22
+Macromolecule #14: Spliceosome-associated protein CWC15 homolog
+Macromolecule #15: RNA helicase aquarius
+Macromolecule #16: SNW domain-containing protein 1
+Macromolecule #17: Peptidyl-prolyl cis-trans isomerase-like 1
+Macromolecule #18: Pleiotropic regulator 1
+Macromolecule #19: Serine/arginine repetitive matrix protein 2
+Macromolecule #20: Pre-mRNA-splicing factor CWC22 homolog
+Macromolecule #21: Pre-mRNA-processing factor 17
+Macromolecule #22: Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16
+Macromolecule #23: Peptidyl-prolyl cis-trans isomerase-like 4
+Macromolecule #24: Pre-mRNA-splicing factor SPF27
+Macromolecule #25: Small nuclear ribonucleoprotein-associated proteins B and B'
+Macromolecule #26: Small nuclear ribonucleoprotein Sm D1
+Macromolecule #27: Small nuclear ribonucleoprotein Sm D2
+Macromolecule #28: Small nuclear ribonucleoprotein F
+Macromolecule #29: Small nuclear ribonucleoprotein E
+Macromolecule #30: Small nuclear ribonucleoprotein G
+Macromolecule #31: Small nuclear ribonucleoprotein Sm D3
+Macromolecule #32: Pre-mRNA-processing factor 19
+Macromolecule #33: Peptidyl-prolyl cis-trans isomerase E
+Macromolecule #34: Splicing factor 3B subunit 1
+Macromolecule #35: Splicing factor 3B subunit 3
+Macromolecule #36: Splicing factor 3B subunit 2
+Macromolecule #37: Splicing factor 3B subunit 4
+Macromolecule #38: PHD finger-like domain-containing protein 5A
+Macromolecule #39: Splicing factor 3B subunit 5
+Macromolecule #40: U2 small nuclear ribonucleoprotein B''
+Macromolecule #41: U2 small nuclear ribonucleoprotein A'
+Macromolecule #2: U5 snRNA
+Macromolecule #5: U6 snRNA
+Macromolecule #6: Pre-mRNA
+Macromolecule #7: U2 snRNA
+Macromolecule #42: INOSITOL HEXAKISPHOSPHATE
+Macromolecule #43: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #44: MAGNESIUM ION
+Macromolecule #45: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 92596 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)