+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse Pendrin bound chloride in inward state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | exchange / transport / slc / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationInorganic anion exchange by SLC26 transporters / iodide transmembrane transporter activity / secondary active sulfate transmembrane transporter activity / monoatomic anion transmembrane transporter activity / regulation of pH / chloride:bicarbonate antiporter activity / animal organ morphogenesis / brush border membrane / regulation of protein localization / apical plasma membrane ...Inorganic anion exchange by SLC26 transporters / iodide transmembrane transporter activity / secondary active sulfate transmembrane transporter activity / monoatomic anion transmembrane transporter activity / regulation of pH / chloride:bicarbonate antiporter activity / animal organ morphogenesis / brush border membrane / regulation of protein localization / apical plasma membrane / extracellular exosome / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Liu QY / Zhang X / Sun L / Chen ZG | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Mouse Pendrin bound chloride in inward state Authors: Liu QY / Zhang X / Sun L / Chen ZG | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35089.map.gz emd_35089.map.gz | 110.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35089-v30.xml emd-35089-v30.xml emd-35089.xml emd-35089.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35089.png emd_35089.png | 124.2 KB | ||
| Filedesc metadata |  emd-35089.cif.gz emd-35089.cif.gz | 5.8 KB | ||
| Others |  emd_35089_half_map_1.map.gz emd_35089_half_map_1.map.gz emd_35089_half_map_2.map.gz emd_35089_half_map_2.map.gz | 110.9 MB 111 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35089 http://ftp.pdbj.org/pub/emdb/structures/EMD-35089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35089 | HTTPS FTP |
-Related structure data
| Related structure data |  8hznMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35089.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35089.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
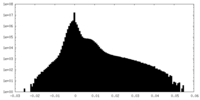
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
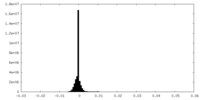
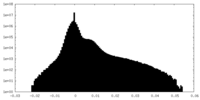
| Density Histograms |
-Half map: #2
| File | emd_35089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
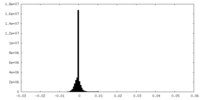
| Density Histograms |
- Sample components
Sample components
-Entire : mouse Pendrin bound chloride in inward state
| Entire | Name: mouse Pendrin bound chloride in inward state |
|---|---|
| Components |
|
-Supramolecule #1: mouse Pendrin bound chloride in inward state
| Supramolecule | Name: mouse Pendrin bound chloride in inward state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Pendrin
| Macromolecule | Name: Pendrin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.769578 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAARGGRSEP PQLAEYSCSY TVSRPVYSEL AFQQQRERRL PERRTLRDSL ARSCSCSRKR AFGVVKTLLP ILDWLPKYRV KEWLLSDII SGVSTGLVGT LQGMAYALLA AVPVQFGLYS AFFPILTYFV FGTSRHISVG PFPVVSLMVG SVVLSMAPDD H FLVPSGNG ...String: MAARGGRSEP PQLAEYSCSY TVSRPVYSEL AFQQQRERRL PERRTLRDSL ARSCSCSRKR AFGVVKTLLP ILDWLPKYRV KEWLLSDII SGVSTGLVGT LQGMAYALLA AVPVQFGLYS AFFPILTYFV FGTSRHISVG PFPVVSLMVG SVVLSMAPDD H FLVPSGNG SALNSTTLDT GTRDAARVLL ASTLTLLVGI IQLVFGGLQI GFIVRYLADP LVGGFTTAAA FQVLVSQLKI VL NVSTKNY NGILSIIYTL IEIFQNIGDT NIADFIAGLL TIIVCMAVKE LNDRFKHRIP VPIPIEVIVT IIATAISYGA NLE KNYNAG IVKSIPSGFL PPVLPSVGLF SDMLAASFSI AVVAYAIAVS VGKVYATKHD YVIDGNQEFI AFGISNVFSG FFSC FVATT ALSRTAVQES TGGKTQVAGL ISAVIVMVAI VALGRLLEPL QKSVLAAVVI ANLKGMFMQV CDVPRLWKQN KTDAV IWVF TCIMSIILGL DLGLLAGLLF ALLTVVLRVQ FPSWNGLGSV PSTDIYKSIT HYKNLEEPEG VKILRFSSPI FYGNVD GFK KCINSTVGFD AIRVYNKRLK ALRRIQKLIK KGQLRATKNG IISDIGSSNN AFEPDEDVEE PEELNIPTKE IEIQVDW NS ELPVKVNVPK VPIHSLVLDC GAVSFLDVVG VRSLRMIVKE FQRIDVNVYF ALLQDDVLEK MEQCGFFDDN IRKDRFFL T VHDAILHLQN QVKSREGQDS LLETVARIRD CKDPLDLMEA EMNAEELDVQ DEAMRRLAS UniProtKB: Pendrin |
-Macromolecule #2: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)