[English] 日本語
 Yorodumi
Yorodumi- EMDB-35080: Far-red light-harvesting complex of Antarctic alga Prasiola crispa -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Prasiola crispa (plant) Prasiola crispa (plant) | ||||||||||||
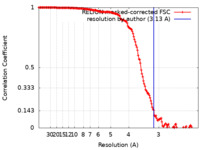
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | ||||||||||||
 Authors Authors | Kosugi M / Kawasaki M / Shibata Y / Moriya T / Adachi N / Senda T | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Uphill energy transfer mechanism for photosynthesis in an Antarctic alga. Authors: Makiko Kosugi / Masato Kawasaki / Yutaka Shibata / Kojiro Hara / Shinichi Takaichi / Toshio Moriya / Naruhiko Adachi / Yasuhiro Kamei / Yasuhiro Kashino / Sakae Kudoh / Hiroyuki Koike / Toshiya Senda /  Abstract: Prasiola crispa, an aerial green alga, forms layered colonies under the severe terrestrial conditions of Antarctica. Since only far-red light is available at a deep layer of the colony, P. crispa has ...Prasiola crispa, an aerial green alga, forms layered colonies under the severe terrestrial conditions of Antarctica. Since only far-red light is available at a deep layer of the colony, P. crispa has evolved a molecular system for photosystem II (PSII) excitation using far-red light with uphill energy transfer. However, the molecular basis underlying this system remains elusive. Here, we purified a light-harvesting chlorophyll (Chl)-binding protein complex from P. crispa (Pc-frLHC) that excites PSII with far-red light and revealed its ring-shaped structure with undecameric 11-fold symmetry at 3.13 Å resolution. The primary structure suggests that Pc-frLHC evolved from LHCI rather than LHCII. The circular arrangement of the Pc-frLHC subunits is unique among eukaryote LHCs and forms unprecedented Chl pentamers at every subunit‒subunit interface near the excitation energy exit sites. The Chl pentamers probably contribute to far-red light absorption. Pc-frLHC's unique Chl arrangement likely promotes PSII excitation with entropy-driven uphill excitation energy transfer. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35080.map.gz emd_35080.map.gz | 55.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35080-v30.xml emd-35080-v30.xml emd-35080.xml emd-35080.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35080_fsc.xml emd_35080_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_35080.png emd_35080.png | 137.8 KB | ||
| Others |  emd_35080_half_map_1.map.gz emd_35080_half_map_1.map.gz emd_35080_half_map_2.map.gz emd_35080_half_map_2.map.gz | 54.8 MB 54.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35080 http://ftp.pdbj.org/pub/emdb/structures/EMD-35080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35080 | HTTPS FTP |
-Validation report
| Summary document |  emd_35080_validation.pdf.gz emd_35080_validation.pdf.gz | 794.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35080_full_validation.pdf.gz emd_35080_full_validation.pdf.gz | 794.3 KB | Display | |
| Data in XML |  emd_35080_validation.xml.gz emd_35080_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_35080_validation.cif.gz emd_35080_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35080 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35080 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35080 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35080 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35080.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35080.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35080_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
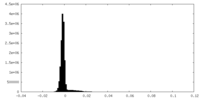
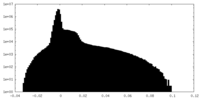
| Density Histograms |
-Half map: #2
| File | emd_35080_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
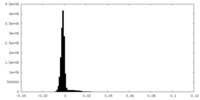
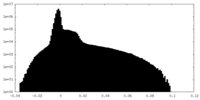
| Density Histograms |
- Sample components
Sample components
-Entire : Far-red light-harvesting complex
| Entire | Name: Far-red light-harvesting complex |
|---|---|
| Components |
|
-Supramolecule #1: Far-red light-harvesting complex
| Supramolecule | Name: Far-red light-harvesting complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Prasiola crispa (plant) Prasiola crispa (plant) |
| Molecular weight | Theoretical: 700 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blot time 5 sec blot force 25. |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1555 / Average exposure time: 64.92 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8hw1: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)