+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hepatitis / HBV / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbile acid:sodium symporter activity / membrane fusion involved in viral entry into host cell / bile acid transmembrane transporter activity / bile acid and bile salt transport / Recycling of bile acids and salts / response to nutrient levels / response to estrogen / cellular response to xenobiotic stimulus / virus receptor activity / basolateral plasma membrane ...bile acid:sodium symporter activity / membrane fusion involved in viral entry into host cell / bile acid transmembrane transporter activity / bile acid and bile salt transport / Recycling of bile acids and salts / response to nutrient levels / response to estrogen / cellular response to xenobiotic stimulus / virus receptor activity / basolateral plasma membrane / response to ethanol / symbiont entry into host cell / virion attachment to host cell / virion membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Hepatitis B virus / Hepatitis B virus /  Ondatra zibethicus (muskrat) Ondatra zibethicus (muskrat) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Asami J / Shimizu T / Ohto U | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of hepatitis B virus receptor binding. Authors: Jinta Asami / Jae-Hyun Park / Yayoi Nomura / Chisa Kobayashi / Junki Mifune / Naito Ishimoto / Tomoko Uemura / Kehong Liu / Yumi Sato / Zhikuan Zhang / Masamichi Muramatsu / Takaji Wakita / ...Authors: Jinta Asami / Jae-Hyun Park / Yayoi Nomura / Chisa Kobayashi / Junki Mifune / Naito Ishimoto / Tomoko Uemura / Kehong Liu / Yumi Sato / Zhikuan Zhang / Masamichi Muramatsu / Takaji Wakita / David Drew / So Iwata / Toshiyuki Shimizu / Koichi Watashi / Sam-Yong Park / Norimichi Nomura / Umeharu Ohto /   Abstract: Hepatitis B virus (HBV), a leading cause of developing hepatocellular carcinoma affecting more than 290 million people worldwide, is an enveloped DNA virus specifically infecting hepatocytes. ...Hepatitis B virus (HBV), a leading cause of developing hepatocellular carcinoma affecting more than 290 million people worldwide, is an enveloped DNA virus specifically infecting hepatocytes. Myristoylated preS1 domain of the HBV large surface protein binds to the host receptor sodium-taurocholate cotransporting polypeptide (NTCP), a hepatocellular bile acid transporter, to initiate viral entry. Here, we report the cryogenic-electron microscopy structure of the myristoylated preS1 (residues 2-48) peptide bound to human NTCP. The unexpectedly folded N-terminal half of the peptide embeds deeply into the outward-facing tunnel of NTCP, whereas the C-terminal half formed extensive contacts on the extracellular surface. Our findings reveal an unprecedented induced-fit mechanism for establishing high-affinity virus-host attachment and provide a blueprint for the rational design of anti-HBV drugs targeting virus entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34982.map.gz emd_34982.map.gz | 23.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34982-v30.xml emd-34982-v30.xml emd-34982.xml emd-34982.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34982.png emd_34982.png | 74.3 KB | ||
| Filedesc metadata |  emd-34982.cif.gz emd-34982.cif.gz | 6 KB | ||
| Others |  emd_34982_half_map_1.map.gz emd_34982_half_map_1.map.gz emd_34982_half_map_2.map.gz emd_34982_half_map_2.map.gz | 23.5 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34982 http://ftp.pdbj.org/pub/emdb/structures/EMD-34982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34982 | HTTPS FTP |
-Related structure data
| Related structure data |  8hryMC  8hrxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34982.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34982.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
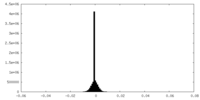
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34982_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
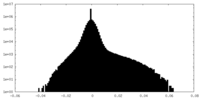
| Density Histograms |
-Half map: #2
| File | emd_34982_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
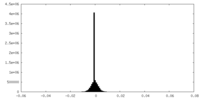
| Density Histograms |
- Sample components
Sample components
-Entire : NTCP-myr-preS1-YN9016Fab
| Entire | Name: NTCP-myr-preS1-YN9016Fab |
|---|---|
| Components |
|
-Supramolecule #1: NTCP-myr-preS1-YN9016Fab
| Supramolecule | Name: NTCP-myr-preS1-YN9016Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium/bile acid cotransporter
| Macromolecule | Name: Sodium/bile acid cotransporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.198883 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEAHNASAPF NFTLPPNFGK RPTDLALSVI LVFMLFFIML SLGCTMEFSK IKAHLWKPKG LAIALVAQYG IMPLTAFVLG KVFRLKNIE ALAILVCGCS PGGNLSNVFS LAMKGDMNLS IVMTTCSTFC ALGMMPLLLY IYSRGIYDGD LKDKVPYKGI V ISLVLVLI ...String: MEAHNASAPF NFTLPPNFGK RPTDLALSVI LVFMLFFIML SLGCTMEFSK IKAHLWKPKG LAIALVAQYG IMPLTAFVLG KVFRLKNIE ALAILVCGCS PGGNLSNVFS LAMKGDMNLS IVMTTCSTFC ALGMMPLLLY IYSRGIYDGD LKDKVPYKGI V ISLVLVLI PCTIGIVLKS KRPQYMRYVI KGGMIIILLC SVAVTVLSAI NVGKSIMFAM TPLLIATSSL MPFIGFLLGY VL SALFCLN GRCRRTVSME TGCQNVQLCS TILNVAFPPE VIGPLFFFPL LYMIFQLGEG LLLIAIFWCY EKFKTPKDKT KMI ENLYFQ GDYKDDDDKH HHHHHHH UniProtKB: Hepatic sodium/bile acid cotransporter |
-Macromolecule #2: Large S protein (Fragment)
| Macromolecule | Name: Large S protein (Fragment) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
| Molecular weight | Theoretical: 6.229594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GTNLSVPNPL GFFPDHQLDP AFKANSENPD WDLNPHKDNW PDANKVGDYK DDDDK UniProtKB: Large S protein |
-Macromolecule #3: Fab heavy chain from antibody IgG clone number YN9016
| Macromolecule | Name: Fab heavy chain from antibody IgG clone number YN9016 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ondatra zibethicus (muskrat) Ondatra zibethicus (muskrat) |
| Molecular weight | Theoretical: 25.306494 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQESGAE LVRPGTSVKM SCKAAGYTFT NYWIGWVKQR PGHGLEWIGD IYPGGGYTNY NEKFKGKATL TADTSSSTAY MQLSSLTSE DSALYYCARM KINNQAWFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS ...String: EVQLQESGAE LVRPGTSVKM SCKAAGYTFT NYWIGWVKQR PGHGLEWIGD IYPGGGYTNY NEKFKGKATL TADTSSSTAY MQLSSLTSE DSALYYCARM KINNQAWFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVPSSTWPSE TVTCNVAHPA SSTKVDKKIV PRDCGCKPCI CTVPEVSS |
-Macromolecule #4: Fab light chain from antibody IgG clone number YN9016
| Macromolecule | Name: Fab light chain from antibody IgG clone number YN9016 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ondatra zibethicus (muskrat) Ondatra zibethicus (muskrat) |
| Molecular weight | Theoretical: 23.667104 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSPAS LSASVGETVT ITCRASENIY SYLTWYQQKQ GKSPQLLVYN AKTLAEGVPS RFSGSGSGTQ FSLKINSLQP EDFGSYYCQ HHYGTPFTFG TGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVMTQSPAS LSASVGETVT ITCRASENIY SYLTWYQQKQ GKSPQLLVYN AKTLAEGVPS RFSGSGSGTQ FSLKINSLQP EDFGSYYCQ HHYGTPFTFG TGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.01% GDN |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.11 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 72000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)