+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GPR21-Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of multicellular organism growth / PKA activation in glucagon signalling / hair follicle placode formation / developmental growth / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway ...positive regulation of multicellular organism growth / PKA activation in glucagon signalling / hair follicle placode formation / developmental growth / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / negative regulation of insulin receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / G protein-coupled receptor activity / bone development / G-protein beta/gamma-subunit complex binding / platelet aggregation / cognition / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / sensory perception of smell / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / insulin receptor signaling pathway / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / glucose homeostasis / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cold-induced thermogenesis / G protein activity / GTPase binding / retina development in camera-type eye / Ca2+ pathway / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / fibroblast proliferation / G alpha (i) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / GTP binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
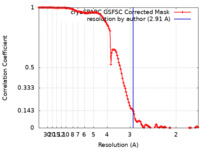
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Wong TS / Gao W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: MedComm (2020) / Year: 2023 Journal: MedComm (2020) / Year: 2023Title: Cryo-EM structure of orphan G protein-coupled receptor GPR21. Authors: Thian-Sze Wong / Wei Gao / Geng Chen / Chen Qiu / Guodong He / Fang Ye / Zhangsong Wu / Zicheng Zeng / Yang Du /  Abstract: GPR21 belongs to class A orphan G protein-coupled receptor (GPCR). The endogenous ligands for human GPR21 remain unidentified. GPR21 expression is associated with developing type 2 diabetes (T2DM), a ...GPR21 belongs to class A orphan G protein-coupled receptor (GPCR). The endogenous ligands for human GPR21 remain unidentified. GPR21 expression is associated with developing type 2 diabetes (T2DM), a multifactorial metabolic disease caused by pancreatic β-cell dysfunction, decreasing insulin production, insulin resistance, and obesity. Animal studies suggested that GPR21 is a potential therapeutic target for T2DM treatment. The underlying mechanisms leading to GPR21 self-activation remain unknown. In our co-expression analysis, we noted that GPR21 could also form a stable complex with an unreported Gα protein subtype, Gαs. To gain further insights into the structural mechanisms of GPR21 activation, we employed cryo-electron microscopy (cryo-EM) and single-particle analysis to resolve the high-resolution structure of GPR21-Gαs complexes. The clear electron density map of the GPR21-Gαs provided direct evidence that GPR21 could couple to Gαs protein at physiological conditions. Thus, GPR21 might mediate previously unexplored pathways in normal or pathological conditions, which warrants further investigation. Structure-guided mutagenesis and biochemical analysis revealed that extracellular loop 2 (ECL2) of GPR21 is essential for the receptor transducing intracellular signal via cAMP. Together, the new structure data reveal a novel signaling cascade of human GPR21 mediated by ECL2 and provide fundamental information for future structure-based drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34903.map.gz emd_34903.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34903-v30.xml emd-34903-v30.xml emd-34903.xml emd-34903.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34903_fsc.xml emd_34903_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34903.png emd_34903.png | 54.5 KB | ||
| Filedesc metadata |  emd-34903.cif.gz emd-34903.cif.gz | 5.9 KB | ||
| Others |  emd_34903_half_map_1.map.gz emd_34903_half_map_1.map.gz emd_34903_half_map_2.map.gz emd_34903_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34903 http://ftp.pdbj.org/pub/emdb/structures/EMD-34903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34903 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34903.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34903.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34903_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
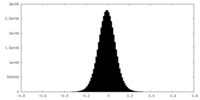
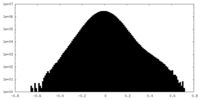
| Density Histograms |
-Half map: #2
| File | emd_34903_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
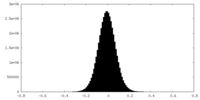
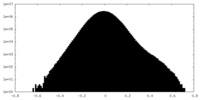
| Density Histograms |
- Sample components
Sample components
-Entire : GPR21-Gs complex
| Entire | Name: GPR21-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: GPR21-Gs complex
| Supramolecule | Name: GPR21-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Probable G-protein coupled receptor 21
| Macromolecule | Name: Probable G-protein coupled receptor 21 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.183879 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LEVLIIVFLT VLIISGNIIV IFVFHCAPLL NHHTTSYFIQ TMAYADLFVG VSCVVPSLSL LHHPLPVEES LTCQIFGFVV SVLKSVSMA SLACISIDRY IAITKPLTYN TLVTPWRLRL CIFLIWLYST LVFLPSFFHW GKPGYHGDVF QWCAESWHTD S YFTLFIVM ...String: LEVLIIVFLT VLIISGNIIV IFVFHCAPLL NHHTTSYFIQ TMAYADLFVG VSCVVPSLSL LHHPLPVEES LTCQIFGFVV SVLKSVSMA SLACISIDRY IAITKPLTYN TLVTPWRLRL CIFLIWLYST LVFLPSFFHW GKPGYHGDVF QWCAESWHTD S YFTLFIVM MLYAPAALIV CFTYFNIFRI CQQHTKDISE RQARFSSQSG ETGEVQACPD KRYAMVLFRI TSVFYILWLP YI IYFLLES STGHSNRFAS FLTTWLAISN SFCNCVIYSL SNSVFQRGLK RL UniProtKB: Probable G-protein coupled receptor 21 |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.747395 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKAT KVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV RACYERSNEY Q LIDCAQYF ...String: DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKAT KVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV RACYERSNEY Q LIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTTGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF NDVTAIIFVV AS SSYNMVI REDNQTNRLQ EALNLFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTTPEDATPE PGE DPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCS VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.198656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD ...String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TFTGHESDIN AICFFPNGNA FA TGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKADRAGVLA GHDNRVSCLG VTD DGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.261229 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TASIAQARKL VEQLKMEANI DRIKVSKAAA DLMAYCEAHA KEDPLLTPVP ASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Nanobody Nb35
| Macromolecule | Name: Nanobody Nb35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.711284 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)