[English] 日本語
 Yorodumi
Yorodumi- EMDB-34836: Complex of GMPCPP microtubule, FAP20 and tubulin (in absence of GTP) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of GMPCPP microtubule, FAP20 and tubulin (in absence of GTP) | |||||||||||||||||||||
 Map data Map data | C1 reconstruction of microtubule:FAP20:tubulin complex | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | doublet microtubule / cilia- and flagella- associated protein 20 (FAP20) / inner junction protein / cilia / STRUCTURAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationaxonemal central pair / axonemal doublet microtubule / positive regulation of cilium-dependent cell motility / establishment of protein localization to organelle / regulation of cilium beat frequency involved in ciliary motility / axoneme assembly / axonemal microtubule / embryonic brain development / positive regulation of axon guidance / microtubule associated complex ...axonemal central pair / axonemal doublet microtubule / positive regulation of cilium-dependent cell motility / establishment of protein localization to organelle / regulation of cilium beat frequency involved in ciliary motility / axoneme assembly / axonemal microtubule / embryonic brain development / positive regulation of axon guidance / microtubule associated complex / motile cilium / cilium assembly / intercellular bridge / modulation of chemical synaptic transmission / cerebral cortex development / structural constituent of cytoskeleton / microtubule cytoskeleton organization / Schaffer collateral - CA1 synapse / neuron migration / mitotic spindle / mitotic cell cycle / microtubule / hydrolase activity / ciliary basal body / protein heterodimerization activity / GTPase activity / GTP binding / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |   | |||||||||||||||||||||
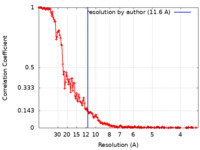
| Method | single particle reconstruction / cryo EM / Resolution: 11.6 Å | |||||||||||||||||||||
 Authors Authors | Bangera M / Sirajuddin M | |||||||||||||||||||||
| Funding support |  India, European Union, 6 items India, European Union, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Doublet microtubule inner junction protein FAP20 recruits tubulin to the microtubule lattice. Authors: Mamata Bangera / Archita Dungdung / Sujana Prabhu / Minhajuddin Sirajuddin /  Abstract: Doublet microtubules of eukaryotic cilia and flagella are made up of a complete A- and an incomplete B-tubule that are fused together. Of the two fusion points, the outer junction is made of ...Doublet microtubules of eukaryotic cilia and flagella are made up of a complete A- and an incomplete B-tubule that are fused together. Of the two fusion points, the outer junction is made of tripartite tubulin connections, while the inner junction contains non-tubulin elements. The latter includes flagellar-associated protein 20 (FAP20) and Parkin co-regulated gene protein (PACRG) that together link the A- and B-tubule at the inner junction. While structures of doublet microtubules reveal molecular details, their assembly is poorly understood. In this study, we purified recombinant FAP20 and characterized its effects on microtubule dynamics. We use in vitro reconstitution and cryo-electron microscopy to show that FAP20 recruits free tubulin to the existing microtubule lattice. Our cryo-electron microscopy reconstruction of microtubule:FAP20:tubulin complex reveals the mode of tubulin recruitment by FAP20 onto microtubules, providing insights into assembly steps of B-tubule closure during doublet microtubule formation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34836.map.gz emd_34836.map.gz | 256.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34836-v30.xml emd-34836-v30.xml emd-34836.xml emd-34836.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34836_fsc.xml emd_34836_fsc.xml | 20.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_34836.png emd_34836.png | 54.9 KB | ||
| Filedesc metadata |  emd-34836.cif.gz emd-34836.cif.gz | 6.2 KB | ||
| Others |  emd_34836_half_map_1.map.gz emd_34836_half_map_1.map.gz emd_34836_half_map_2.map.gz emd_34836_half_map_2.map.gz | 257.8 MB 258.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34836 http://ftp.pdbj.org/pub/emdb/structures/EMD-34836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34836 | HTTPS FTP |
-Validation report
| Summary document |  emd_34836_validation.pdf.gz emd_34836_validation.pdf.gz | 953 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34836_full_validation.pdf.gz emd_34836_full_validation.pdf.gz | 952.6 KB | Display | |
| Data in XML |  emd_34836_validation.xml.gz emd_34836_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_34836_validation.cif.gz emd_34836_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34836 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34836 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34836.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34836.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C1 reconstruction of microtubule:FAP20:tubulin complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.78 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1 for C1 reconstruction of microtubule:FAP20:tubulin complex
| File | emd_34836_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for C1 reconstruction of microtubule:FAP20:tubulin complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 for C1 reconstruction of microtubule:FAP20:tubulin complex
| File | emd_34836_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for C1 reconstruction of microtubule:FAP20:tubulin complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of GMPCPP microtubule with FAP20 and tubulin in absence of GTP
| Entire | Name: Complex of GMPCPP microtubule with FAP20 and tubulin in absence of GTP |
|---|---|
| Components |
|
-Supramolecule #1: Complex of GMPCPP microtubule with FAP20 and tubulin in absence of GTP
| Supramolecule | Name: Complex of GMPCPP microtubule with FAP20 and tubulin in absence of GTP type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: tublin
| Supramolecule | Name: tublin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Cilia- and flagella-associated protein 20
| Supramolecule | Name: Cilia- and flagella-associated protein 20 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin alpha chain
| Macromolecule | Name: Tubulin alpha chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS IYPAPQVSTA VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL ISQIVSSITA SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN QMVKCDPRHG KYMACCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDMAALEK DYEEVGVDSV EGEGEEEGEE Y UniProtKB: Tubulin alpha chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVM ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVM PSPKVSDTVV EPYNATLSVH QLVENTDETY CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDSKNMM AACDPRHGRY LTVAAIFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK TAVCDIPPRG LKMSATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA DEQGEFEEEE GEDEA UniProtKB: Tubulin beta chain |
-Macromolecule #3: Cilia- and flagella-associated protein 20
| Macromolecule | Name: Cilia- and flagella-associated protein 20 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MFKNAFQSGF LSVLYSIGSK PLEIWDKQVS NGHIKRITDA DIQSSVLEIM GQNVSTTYIT CPADPNKTLG IKLPFLVLII KNLNKYFSFE VQVLDDKNVR RRFRASNYQS TTRVKPFICT MPMRLDSGWN QIQFNLSDFT RRAYGTNYIE TLRVQVHANC RIRRIYFSDR ...String: MFKNAFQSGF LSVLYSIGSK PLEIWDKQVS NGHIKRITDA DIQSSVLEIM GQNVSTTYIT CPADPNKTLG IKLPFLVLII KNLNKYFSFE VQVLDDKNVR RRFRASNYQS TTRVKPFICT MPMRLDSGWN QIQFNLSDFT RRAYGTNYIE TLRVQVHANC RIRRIYFSDR LYSEEELPAE FKLFLPIQKS LEVLFQGPGS HHHHHHHHH UniProtKB: Cilia- and flagella-associated protein 20 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Details: BRB80 buffer : 80mM PIPES, 2mM MgCl2, 1mM EGTA supplemented with 1mM GMPCPP |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.025 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: GMPCPP MT seeds in warm BRB80 was applied to the grid followed by second application of pre-warmed solution containing FAP20 and tubulin. After an incubation time of 1 minute, grid was ...Details: GMPCPP MT seeds in warm BRB80 was applied to the grid followed by second application of pre-warmed solution containing FAP20 and tubulin. After an incubation time of 1 minute, grid was blotted for 3 seconds before plunging into liquid ethane.. |
| Details | Goat brain tubulin was polymerized with 1mM GMPCPP at 310K for 2 hours, spun on warm 50 percent BRB80 sucrose cushion and resuspended in warm BRB80 buffer. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1467 / Average exposure time: 2.0 sec. / Average electron dose: 39.55 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)