+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Gq-coupled MRGPRX1 bound with Compound-16 | |||||||||
 Map data Map data | CryoEM structure of Gq coupled MRGPRX1 binding with compound 16 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to chloroquine / acute-phase response / electron transport chain / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway ...response to chloroquine / acute-phase response / electron transport chain / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / transmembrane signaling receptor activity / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / electron transfer activity / periplasmic space / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / protein-containing complex binding / cell surface / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Gan B / Yu LY / Ren RB | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Mechanism of agonist-induced activation of the human itch receptor MRGPRX1. Authors: Bing Gan / Leiye Yu / Haifeng Yang / Haizhan Jiao / Bin Pang / Yian Chen / Chen Wang / Rui Lv / Hongli Hu / Zhijian Cao / Ruobing Ren /  Abstract: Mas-related G-protein-coupled receptors X1-X4 (MRGPRX1-X4) are 4 primate-specific receptors that are recently reported to be responsible for many biological processes, including itch sensation, pain ...Mas-related G-protein-coupled receptors X1-X4 (MRGPRX1-X4) are 4 primate-specific receptors that are recently reported to be responsible for many biological processes, including itch sensation, pain transmission, and inflammatory reactions. MRGPRX1 is the first identified human MRGPR, and its expression is restricted to primary sensory neurons. Due to its dual roles in itch and pain signaling pathways, MRGPRX1 has been regarded as a promising target for itch remission and pain inhibition. Here, we reported a cryo-electron microscopy (cryo-EM) structure of Gq-coupled MRGPRX1 in complex with a synthetic agonist compound 16 in an active conformation at an overall resolution of 3.0 Å via a NanoBiT tethering strategy. Compound 16 is a new pain-relieving compound with high potency and selectivity to MRGPRX1 over other MRGPRXs and opioid receptor. MRGPRX1 was revealed to share common structural features of the Gq-mediated receptor activation mechanism of MRGPRX family members, but the variable residues in orthosteric pocket of MRGPRX1 exhibit the unique agonist recognition pattern, potentially facilitating to design MRGPRX1-specific modulators. Together with receptor activation and itch behavior evaluation assays, our study provides a structural snapshot to modify therapeutic molecules for itch relieving and analgesia targeting MRGPRX1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34833.map.gz emd_34833.map.gz | 85.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34833-v30.xml emd-34833-v30.xml emd-34833.xml emd-34833.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34833.png emd_34833.png | 92.6 KB | ||
| Filedesc metadata |  emd-34833.cif.gz emd-34833.cif.gz | 7 KB | ||
| Others |  emd_34833_half_map_1.map.gz emd_34833_half_map_1.map.gz emd_34833_half_map_2.map.gz emd_34833_half_map_2.map.gz | 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34833 http://ftp.pdbj.org/pub/emdb/structures/EMD-34833 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34833 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34833 | HTTPS FTP |
-Validation report
| Summary document |  emd_34833_validation.pdf.gz emd_34833_validation.pdf.gz | 943.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34833_full_validation.pdf.gz emd_34833_full_validation.pdf.gz | 943 KB | Display | |
| Data in XML |  emd_34833_validation.xml.gz emd_34833_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_34833_validation.cif.gz emd_34833_validation.cif.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34833 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34833 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34833 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34833 | HTTPS FTP |
-Related structure data
| Related structure data |  8hj5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34833.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34833.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of Gq coupled MRGPRX1 binding with compound 16 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
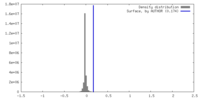
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half A map
| File | emd_34833_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half B map
| File | emd_34833_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gq coupled MRGPRX1 bound with compound 16
| Entire | Name: Gq coupled MRGPRX1 bound with compound 16 |
|---|---|
| Components |
|
-Supramolecule #1: Gq coupled MRGPRX1 bound with compound 16
| Supramolecule | Name: Gq coupled MRGPRX1 bound with compound 16 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.855578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY ...String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY IPTQQDVLRT RVKTSGIFET KFQVDKVNFH MFDVGAQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL ND FKSIWNN RWLRTISVIL FLNKQDLLAE KVLAGKSKIE DYFPEFARYT TPEDATPEPG EDPRVTRAKY FIRKEFVDIS TAS GDGRHI CYPHFTCSVD TENARRIFND CKDIILQMNL REYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.005895 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH LEVLFQGPGS SGSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSR LLVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE L AGHTGYLS ...String: HHHHHHHHHH LEVLFQGPGS SGSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSR LLVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE L AGHTGYLS CCRFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CR QTFTGHE SDINAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDA LKADRA GVLAGHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGASGA SGASGASVSG WRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Macromolecule #5: Soluble cytochrome b562,Mas-related G-protein coupled receptor me...
| Macromolecule | Name: Soluble cytochrome b562,Mas-related G-protein coupled receptor member X1,LgBit type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.462594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKLEVLFQ GPGSADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMDPTISTLD TELTPINGTE E TLCYKQTL ...String: MKTIIALSYI FCLVFADYKD DDDKLEVLFQ GPGSADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMDPTISTLD TELTPINGTE E TLCYKQTL SLTVLTCIVS LVGLTGNAVV LWLLGCRMRR NAFSIYILNL AAADFLFLSG RLIYSLLSFI SIPHTISKIL YP VMMFSYF AGLSFLSAVS TERCLSVLWP IWYRCHRPTH LSAVVCVLLW ALSLLRSILE WMLCGFLFSG ADSAWCQTSD FIT VAWLIF LCVVLCGSSL VLLIRILCGS RKIPLTRLYV TILLTVLVFL LCGLPFGIQF FLFLWIHVDR EVLFCHVHLV SIFL SALNS SANPIIYFFV GSFRQRQNRQ NLKLVLQRAL QDASEVDEGG GQLPEEILEL SGSRLEQGSA GSVFTLEDFV GDWEQ TAAY NLDQVLEQGG VSSLLQNLAV SVTPIQRIVR SGENALKIDI HVIIPYEGLS ADQMAQIEEV FKVVYPVDDH HFKVIL PYG TLVIDGVTPN MLNYFGRPYE GIAVFDGKKI TVTGTLWNGN KIIDERLITP DGSMLFRVTI NSLEHHHHHH HHHH UniProtKB: Soluble cytochrome b562, Mas-related G-protein coupled receptor member X1 |
-Macromolecule #6: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.057271 KDa |
| Recombinant expression | Organism: Bacteria (eubacteria) |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMQVQLQES GGGLVQPGGS LRLSCAASGF TFSNYKMNWV RQAPGKGLEW VSDISQSGAS ISYTGSVKG RFTISRDNAK NTLYLQMNSL KPEDTAVYYC ARCPAPFTRD CFDVTSTTYA YRGQGTQVTV SSHHHHHH |
-Macromolecule #7: N-{2-[(1-aminoisoquinolin-6-yl)oxy]-4-methylphenyl}-2-methoxybenz...
| Macromolecule | Name: N-{2-[(1-aminoisoquinolin-6-yl)oxy]-4-methylphenyl}-2-methoxybenzene-1-sulfonamide type: ligand / ID: 7 / Number of copies: 1 / Formula: U2U |
|---|---|
| Molecular weight | Theoretical: 435.496 Da |
| Chemical component information |  ChemComp-U2U: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.15 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1006848 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)