[English] 日本語
 Yorodumi
Yorodumi- EMDB-34821: A cryo-EM structure of B. oleracea RNA polymerase V elongation co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA-dependent RNA polymerase V / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase IV complex / RNA polymerase V complex / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / RNA polymerase I complex / RNA polymerase III complex / tRNA transcription by RNA polymerase III / RNA polymerase II, core complex / transcription by RNA polymerase I / transcription-coupled nucleotide-excision repair / transcription initiation at RNA polymerase II promoter ...RNA polymerase IV complex / RNA polymerase V complex / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / RNA polymerase I complex / RNA polymerase III complex / tRNA transcription by RNA polymerase III / RNA polymerase II, core complex / transcription by RNA polymerase I / transcription-coupled nucleotide-excision repair / transcription initiation at RNA polymerase II promoter / DNA-directed RNA polymerase activity / nucleic acid binding / transcription by RNA polymerase II / protein dimerization activity / DNA-templated transcription / nucleolus / DNA binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Hu H / Xie G / Du X / Du J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structure and mechanism of the plant RNA polymerase V. Authors: Guohui Xie / Xuan Du / Hongmiao Hu / Sisi Li / Xiaofeng Cao / Steven E Jacobsen / Jiamu Du /   Abstract: In addition to the conserved RNA polymerases I to III (Pols I to III) in eukaryotes, two atypical polymerases, Pols IV and V, specifically produce noncoding RNA in the RNA-directed DNA methylation ...In addition to the conserved RNA polymerases I to III (Pols I to III) in eukaryotes, two atypical polymerases, Pols IV and V, specifically produce noncoding RNA in the RNA-directed DNA methylation pathway in plants. Here, we report on the structures of cauliflower Pol V in the free and elongation conformations. A conserved tyrosine residue of NRPE2 stacks with a double-stranded DNA branch of the transcription bubble to potentially attenuate elongation by inducing transcription stalling. The nontemplate DNA strand is captured by NRPE2 to enhance backtracking, thereby increasing 3'-5' cleavage, which likely underpins Pol V's high fidelity. The structures also illuminate the mechanism of Pol V transcription stalling and enhanced backtracking, which may be important for Pol V's retention on chromatin to serve its function in tethering downstream factors for RNA-directed DNA methylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34821.map.gz emd_34821.map.gz | 96 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34821-v30.xml emd-34821-v30.xml emd-34821.xml emd-34821.xml | 32.5 KB 32.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34821.png emd_34821.png | 137.3 KB | ||
| Filedesc metadata |  emd-34821.cif.gz emd-34821.cif.gz | 9.1 KB | ||
| Others |  emd_34821_half_map_1.map.gz emd_34821_half_map_1.map.gz emd_34821_half_map_2.map.gz emd_34821_half_map_2.map.gz | 81.1 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34821 http://ftp.pdbj.org/pub/emdb/structures/EMD-34821 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34821 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34821 | HTTPS FTP |
-Related structure data
| Related structure data |  8himMC  8hilC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34821.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34821.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.095 Å | ||||||||||||||||||||||||||||||||||||
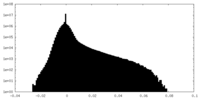
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34821_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
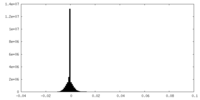
| Density Histograms |
-Half map: #1
| File | emd_34821_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
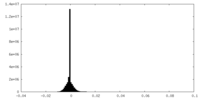
| Density Histograms |
- Sample components
Sample components
+Entire : DNA-directed RNA polymerase V elongation complex
+Supramolecule #1: DNA-directed RNA polymerase V elongation complex
+Supramolecule #2: DNA-directed RNA polymerase
+Supramolecule #3: DNA-RNA
+Macromolecule #1: DNA (34-MER)
+Macromolecule #2: DNA (34-MER)
+Macromolecule #3: RNA (5'-R(*UP*AP*UP*AP*UP*GP*CP*AP*UP*AP*AP*AP*GP*AP*CP*CP*AP*GP*...
+Macromolecule #4: DNA-directed RNA polymerase V largest subunit
+Macromolecule #5: RPOLD domain-containing protein
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #8: RNA_pol_L_2 domain-containing protein
+Macromolecule #9: DNA-directed RNA polymerases II, IV and V subunit 12
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #11: DNA-directed RNA polymerase subunit
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #13: DNA-directed RNA polymerase IV and V subunit 2
+Macromolecule #14: MAGNESIUM ION
+Macromolecule #15: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 8 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 280 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 5694 / Average exposure time: 2.9975 sec. / Average electron dose: 1.5625 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8him: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)