[English] 日本語
 Yorodumi
Yorodumi- EMDB-34809: Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | CRISPR / cas / adaptation / IMMUNE SYSTEM | |||||||||||||||||||||||||||
| Biological species |  Streptococcus thermophilus DGCC 7710 (bacteria) / Streptococcus thermophilus DGCC 7710 (bacteria) /  Phage #D (virus) Phage #D (virus) | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||||||||||||||||||||
 Authors Authors | Chen Q / Luo Y | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Innovation (Camb) / Year: 2023 Journal: Innovation (Camb) / Year: 2023Title: DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism. Authors: Dongmei Tang / Tingting Jia / Yongbo Luo / Biqin Mou / Jie Cheng / Shiqian Qi / Shaohua Yao / Zhaoming Su / Yamei Yu / Qiang Chen /  Abstract: In the spacer acquisition stage of CRISPR-Cas immunity, spacer orientation and protospacer adjacent motif (PAM) removal are two prerequisites for functional spacer integration. Cas4 has been ...In the spacer acquisition stage of CRISPR-Cas immunity, spacer orientation and protospacer adjacent motif (PAM) removal are two prerequisites for functional spacer integration. Cas4 has been implicated in both processing the prespacer and determining the spacer orientation. In Cas4-lacking systems, host 3'-5' DnaQ family exonucleases were recently reported to play a Cas4-like role. However, the molecular details of DnaQ functions remain elusive. Here, we characterized the spacer acquisition of the adaptation module of the type I-E system, in which a DnaQ domain naturally fuses with Cas2. We presented X-ray crystal structures and cryo-electron microscopy structures of this adaptation module. Our biochemical data showed that DnaQ trimmed PAM-containing and PAM-deficient overhangs with different efficiencies. Based on these results, we proposed a time-dependent model for DnaQ-mediated spacer acquisition to elucidate PAM removal and spacer orientation determination in Cas4-lacking CRISPR-Cas systems. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34809.map.gz emd_34809.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34809-v30.xml emd-34809-v30.xml emd-34809.xml emd-34809.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34809.png emd_34809.png | 104.9 KB | ||
| Filedesc metadata |  emd-34809.cif.gz emd-34809.cif.gz | 6.5 KB | ||
| Others |  emd_34809_half_map_1.map.gz emd_34809_half_map_1.map.gz emd_34809_half_map_2.map.gz emd_34809_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34809 http://ftp.pdbj.org/pub/emdb/structures/EMD-34809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34809 | HTTPS FTP |
-Validation report
| Summary document |  emd_34809_validation.pdf.gz emd_34809_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34809_full_validation.pdf.gz emd_34809_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_34809_validation.xml.gz emd_34809_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_34809_validation.cif.gz emd_34809_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34809 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34809 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34809 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34809 | HTTPS FTP |
-Related structure data
| Related structure data |  8hi1MC  8h18C  8h2fC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34809.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34809.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34809_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
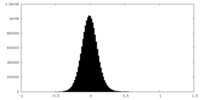
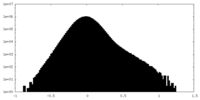
| Density Histograms |
-Half map: #2
| File | emd_34809_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
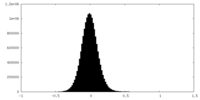
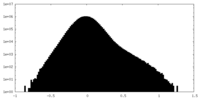
| Density Histograms |
- Sample components
Sample components
-Entire : Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex
| Entire | Name: Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex
| Supramolecule | Name: Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
-Macromolecule #1: CRISPR-associated endonuclease Cas1
| Macromolecule | Name: CRISPR-associated endonuclease Cas1 / type: protein_or_peptide / ID: 1 / Details: WP_024704118.1 / Number of copies: 4 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
| Molecular weight | Theoretical: 35.54957 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GASGSMVEKN GAKKTSLREL PKISDRVSFI YVEHAKINRV DSAITVLDSR GTVRIPAAMI GVLLLGPGTD ISHRAVELIG DTGTSMVWV GERGVRQYAH GRSLAHSTKF LEKQAKLVSN SRLRLAVARK MYQMRFPDED VSAMTMQQLR GREGARVRRV Y RLQSEKYQ ...String: GASGSMVEKN GAKKTSLREL PKISDRVSFI YVEHAKINRV DSAITVLDSR GTVRIPAAMI GVLLLGPGTD ISHRAVELIG DTGTSMVWV GERGVRQYAH GRSLAHSTKF LEKQAKLVSN SRLRLAVARK MYQMRFPDED VSAMTMQQLR GREGARVRRV Y RLQSEKYQ VSWTKREYNP DDFEGGDIVN QALSAANVAL YGLVHSIVIA LGASPGLGFV HTGHDLSFIY DIADLYKAEL TI PLAFEIA ANFTEIDDIG KIARQKVRDS FVDGKLIVRI VQDIQYLFDL DDDEELLVDT LSLWDDKDML VKHGVSYKEE L |
-Macromolecule #2: Type I-E CRISPR-associated endoribonuclease Cas2
| Macromolecule | Name: Type I-E CRISPR-associated endoribonuclease Cas2 / type: protein_or_peptide / ID: 2 / Details: WP_024704117.1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
| Molecular weight | Theoretical: 13.94086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPFTVVTLKS VPPSLRGDLT KWMQEIAIGV YVGNFNSRIR EKLWNRIQAN VGEGEATISY YYRNEIGYQF DMINSQKSVV DFDGIPLVL IPNSKTSSEN YPKLGYSNAA KSRKIKRYSS YRG |
-Macromolecule #3: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Phage #D (virus) Phage #D (virus) |
| Molecular weight | Theoretical: 11.741589 KDa |
| Sequence | String: (DA)(DA)(DA)(DC)(DA)(DC)(DC)(DA)(DG)(DA) (DA)(DC)(DG)(DA)(DG)(DT)(DA)(DG)(DT)(DA) (DA)(DA)(DT)(DT)(DG)(DA)(DT)(DG)(DT) (DT)(DG)(DT)(DC)(DT)(DT)(DG)(DC)(DT) |
-Macromolecule #4: DNA (31-MER)
| Macromolecule | Name: DNA (31-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Phage #D (virus) Phage #D (virus) |
| Molecular weight | Theoretical: 11.708511 KDa |
| Sequence | String: (DA)(DT)(DT)(DT)(DA)(DC)(DT)(DA)(DC)(DT) (DC)(DG)(DT)(DT)(DC)(DT)(DG)(DG)(DT)(DG) (DT)(DT)(DT)(DT)(DT)(DG)(DT)(DG)(DT) (DT)(DT)(DA)(DA)(DT)(DG)(DA)(DT)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.741 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.465 µm / Nominal defocus min: 1.213 µm |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)