+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human SGLT2-MAP17 complex with Sotagliflozin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion transport / Sodium transport / Sugar transport / Symport / Transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlow-affinity D-glucose:sodium symporter activity / Defective SLC5A2 causes renal glucosuria (GLYS1) / alpha-glucoside transport / alpha-glucoside transmembrane transporter activity / D-glucose:sodium symporter activity / hexose transmembrane transport / renal D-glucose absorption / D-glucose import across plasma membrane / Cellular hexose transport / D-glucose transmembrane transporter activity ...low-affinity D-glucose:sodium symporter activity / Defective SLC5A2 causes renal glucosuria (GLYS1) / alpha-glucoside transport / alpha-glucoside transmembrane transporter activity / D-glucose:sodium symporter activity / hexose transmembrane transport / renal D-glucose absorption / D-glucose import across plasma membrane / Cellular hexose transport / D-glucose transmembrane transporter activity / sodium ion import across plasma membrane / sodium ion transport / carbohydrate metabolic process / apical plasma membrane / extracellular exosome / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
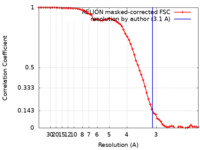
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Hiraizumi M / Kishida H / Miyaguchi I / Nureki O | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter. Authors: Masahiro Hiraizumi / Tomoya Akashi / Kouta Murasaki / Hiroyuki Kishida / Taichi Kumanomidou / Nao Torimoto / Osamu Nureki / Ikuko Miyaguchi /  Abstract: Sodium-glucose cotransporter 2 (SGLT2) is imporant in glucose reabsorption. SGLT2 inhibitors suppress renal glucose reabsorption, therefore reducing blood glucose levels in patients with type 2 ...Sodium-glucose cotransporter 2 (SGLT2) is imporant in glucose reabsorption. SGLT2 inhibitors suppress renal glucose reabsorption, therefore reducing blood glucose levels in patients with type 2 diabetes. We and others have developed several SGLT2 inhibitors starting from phlorizin, a natural product. Using cryo-electron microscopy, we present the structures of human (h)SGLT2-MAP17 complexed with five natural or synthetic inhibitors. The four synthetic inhibitors (including canagliflozin) bind the transporter in the outward conformations, while phlorizin binds it in the inward conformation. The phlorizin-hSGLT2 interaction exhibits biphasic kinetics, suggesting that phlorizin alternately binds to the extracellular and intracellular sides. The Na-bound outward-facing and unbound inward-open structures of hSGLT2-MAP17 suggest that the MAP17-associated bundle domain functions as a scaffold, with the hash domain rotating around the Na-binding site. Thus, Na binding stabilizes the outward-facing conformation, and its release promotes state transition to inward-open conformation, exhibiting a role of Na in symport mechanism. These results provide structural evidence for the Na-coupled alternating-access mechanism proposed for the transporter family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34737.map.gz emd_34737.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34737-v30.xml emd-34737-v30.xml emd-34737.xml emd-34737.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34737_fsc.xml emd_34737_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_34737.png emd_34737.png | 184.2 KB | ||
| Masks |  emd_34737_msk_1.map emd_34737_msk_1.map | 6.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34737.cif.gz emd-34737.cif.gz | 6.2 KB | ||
| Others |  emd_34737_half_map_1.map.gz emd_34737_half_map_1.map.gz emd_34737_half_map_2.map.gz emd_34737_half_map_2.map.gz | 6.1 MB 6.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34737 http://ftp.pdbj.org/pub/emdb/structures/EMD-34737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34737 | HTTPS FTP |
-Validation report
| Summary document |  emd_34737_validation.pdf.gz emd_34737_validation.pdf.gz | 796.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34737_full_validation.pdf.gz emd_34737_full_validation.pdf.gz | 796 KB | Display | |
| Data in XML |  emd_34737_validation.xml.gz emd_34737_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_34737_validation.cif.gz emd_34737_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34737 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34737 | HTTPS FTP |
-Related structure data
| Related structure data |  8hg7MC  8hb0C  8hdhC  8hezC  8hinC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34737.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34737.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.10667 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34737_msk_1.map emd_34737_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of Sodium/glucose cotransporter 2 with PDZK1-inter...
| Entire | Name: Binary complex of Sodium/glucose cotransporter 2 with PDZK1-interacting protein 1. |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of Sodium/glucose cotransporter 2 with PDZK1-inter...
| Supramolecule | Name: Binary complex of Sodium/glucose cotransporter 2 with PDZK1-interacting protein 1. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Electrogenic Na+-coupled sugar simporter that actively transports D-glucose at the plasma membrane, with a Na+ to sugar coupling ratio of 1:1. Transporter activity is driven by a ...Details: Electrogenic Na+-coupled sugar simporter that actively transports D-glucose at the plasma membrane, with a Na+ to sugar coupling ratio of 1:1. Transporter activity is driven by a transmembrane Na+ electrochemical gradient set by the Na+/K+ pump |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium/glucose cotransporter 2
| Macromolecule | Name: Sodium/glucose cotransporter 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.247703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGSMEEHTE AGSAPEMGAQ KALIDNPADI LVIAAYFLLV IGVGLWSMCR TNRGTVGGYF LAGRSMVWWP VGASLFASNI GSGHFVGLA GTGAASGLAV AGFEWNALFV VLLLGWLFAP VYLTAGVITM PQYLRKRFGG RRIRLYLSVL SLFLYIFTKI S VDMFSGAV ...String: GPGSMEEHTE AGSAPEMGAQ KALIDNPADI LVIAAYFLLV IGVGLWSMCR TNRGTVGGYF LAGRSMVWWP VGASLFASNI GSGHFVGLA GTGAASGLAV AGFEWNALFV VLLLGWLFAP VYLTAGVITM PQYLRKRFGG RRIRLYLSVL SLFLYIFTKI S VDMFSGAV FIQQALGWNI YASVIALLGI TMIYTVTGGL AALMYTDTVQ TFVILGGACI LMGYAFHEVG GYSGLFDKYL GA ATSLTVS EDPAVGNISS FCYRPRPDSY HLLRHPVTGD LPWPALLLGL TIVSGWYWCS DQVIVQRCLA GKSLTHIKAG CIL CGYLKL TPMFLMVMPG MISRILYPDE VACVVPEVCR RVCGTEVGCS NIAYPRLVVK LMPNGLRGLM LAVMLAALMS SLAS IFNSS STLFTMDIYT RLRPRAGDRE LLLVGRLWVV FIVVVSVAWL PVVQAAQGGQ LFDYIQAVSS YLAPPVSAVF VLALF VPRV NEQGAFWGLI GGLLMGLARL IPEFSFGSGS CVQPSACPAF LCGVHYLYFA IVLFFCSGLL TLTVSLCTAP IPRKHL HRL VFSLRHSKEE REDLDADEQQ GSSLPVQNGC PESAMEMNEP QAPAPSLFRQ CLLWFCGMSR GGVGSPPPLT QEEAAAA AR RLEDISEDPS WARVVNLNAL LMMAVAVFLW GFYA UniProtKB: Sodium/glucose cotransporter 2 |
-Macromolecule #2: PDZK1-interacting protein 1
| Macromolecule | Name: PDZK1-interacting protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.235 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSALSLLILG LLTAVPPASC QQGLGNLQPW MQGLIAVAVF LVLVAIAFAV NHFWCQEEPE PAHMILTVGN KADGVLVGTD GRYSSMAAS FRSSEHENAY ENVPEEEGKV RSTPM UniProtKB: PDZK1-interacting protein 1 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]...
| Macromolecule | Name: (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]-6-methylsulfanyl-oxane-3,4,5-triol type: ligand / ID: 4 / Number of copies: 1 / Formula: LFL |
|---|---|
| Molecular weight | Theoretical: 424.938 Da |
| Chemical component information | 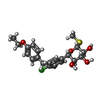 ChemComp-LFL: |
-Macromolecule #5: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 5 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)