+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribonuclease / DOUBLE STRANDED RNA / RNA BINDING PROTEIN-RNA COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationfollicle cell of egg chamber stalk formation / positive regulation of Toll signaling pathway / lncRNA catabolic process / MicroRNA (miRNA) biogenesis / RNAi-mediated antiviral immune response / Small interfering RNA (siRNA) biogenesis / PKR-mediated signaling / female germ-line stem cell asymmetric division / regulation of regulatory ncRNA processing / dosage compensation by hyperactivation of X chromosome ...follicle cell of egg chamber stalk formation / positive regulation of Toll signaling pathway / lncRNA catabolic process / MicroRNA (miRNA) biogenesis / RNAi-mediated antiviral immune response / Small interfering RNA (siRNA) biogenesis / PKR-mediated signaling / female germ-line stem cell asymmetric division / regulation of regulatory ncRNA processing / dosage compensation by hyperactivation of X chromosome / RISC complex binding / global gene silencing by mRNA cleavage / apoptotic DNA fragmentation / germ-line stem cell population maintenance / ribonuclease III / deoxyribonuclease I activity / detection of virus / RISC-loading complex / miRNA metabolic process / RISC complex assembly / regulatory ncRNA-mediated post-transcriptional gene silencing / ribonuclease III activity / pre-miRNA processing / siRNA processing / siRNA binding / ATP-dependent activity, acting on RNA / RISC complex / positive regulation of innate immune response / positive regulation of defense response to virus by host / central nervous system development / mRNA 3'-UTR binding / locomotory behavior / helicase activity / cellular response to virus / cytoplasmic ribonucleoprotein granule / heterochromatin formation / double-stranded RNA binding / defense response to virus / perinuclear region of cytoplasm / ATP hydrolysis activity / RNA binding / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.72 Å | |||||||||
 Authors Authors | Su S / Wang J / Wang HW / Ma J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers. Authors: Ting Deng / Shichen Su / Xun Yuan / Jinqiu He / Ying Huang / Jinbiao Ma / Jia Wang /  Abstract: Small interference RNAs are the key components of RNA interference, a conserved RNA silencing or viral defense mechanism in many eukaryotes. In Drosophila melanogaster, Dicer-2 (DmDcr-2)-mediated ...Small interference RNAs are the key components of RNA interference, a conserved RNA silencing or viral defense mechanism in many eukaryotes. In Drosophila melanogaster, Dicer-2 (DmDcr-2)-mediated RNAi pathway plays important roles in defending against viral infections and protecting genome integrity. During the maturation of siRNAs, two cofactors can regulate DmDcr-2's functions: Loqs-PD that is required for dsRNA processing, and R2D2 that is essential for the subsequent loading of siRNAs into effector Ago2 to form RISC complexes. However, due to the lack of structural information, it is still unclear whether R2D2 and Loqs-PD affect the functions of DmDcr-2 simultaneously. Here we present several cryo-EM structures of DmDcr-2/R2D2/Loqs-PD complex bound to dsRNAs with various lengths by the Helicase domain. These structures revealed that R2D2 and Loqs-PD can bind to different regions of DmDcr-2 without interfering with each other. Furthermore, the cryo-EM results demonstrate that these complexes can form large oligomers and assemble into fibers. The formation and depolymerization of these oligomers are associated with ATP hydrolysis. These findings provide insights into the structural mechanism of DmDcr-2 and its cofactors during siRNA processing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34707.map.gz emd_34707.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34707-v30.xml emd-34707-v30.xml emd-34707.xml emd-34707.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34707.png emd_34707.png | 91.8 KB | ||
| Others |  emd_34707_half_map_1.map.gz emd_34707_half_map_1.map.gz emd_34707_half_map_2.map.gz emd_34707_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34707 http://ftp.pdbj.org/pub/emdb/structures/EMD-34707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34707 | HTTPS FTP |
-Related structure data
| Related structure data |  8hf0MC  8hf1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34707.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34707.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0742 Å | ||||||||||||||||||||||||||||||||||||
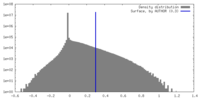
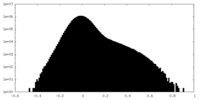
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34707_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
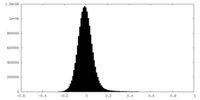
| Density Histograms |
-Half map: #1
| File | emd_34707_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
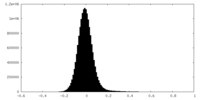
| Density Histograms |
- Sample components
Sample components
-Entire : DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state
| Entire | Name: DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state |
|---|---|
| Components |
|
-Supramolecule #1: DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state
| Supramolecule | Name: DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Dicer-2, isoform A
| Macromolecule | Name: Dicer-2, isoform A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: deoxyribonuclease I |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 198.006688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDVEIKPRG YQLRLVDHLT KSNGIVYLPT GSGKTFVAIL VLKRFSQDFD KPIESGGKRA LFMCNTVELA RQQAMAVRRC TNFKVGFYV GEQGVDDWTR GMWSDEIKKN QVLVGTAQVF LDMVTQTYVA LSSLSVVIID ECHHGTGHHP FREFMRLFTI A NQTKLPRV ...String: MEDVEIKPRG YQLRLVDHLT KSNGIVYLPT GSGKTFVAIL VLKRFSQDFD KPIESGGKRA LFMCNTVELA RQQAMAVRRC TNFKVGFYV GEQGVDDWTR GMWSDEIKKN QVLVGTAQVF LDMVTQTYVA LSSLSVVIID ECHHGTGHHP FREFMRLFTI A NQTKLPRV VGLTGVLIKG NEITNVATKL KELEITYRGN IITVSDTKEM ENVMLYATKP TEVMVSFPHQ EQVLTVTRLI SA EIEKFYV SLDLMNIGVQ PIRRSKSLQC LRDPSKKSFV KQLFNDFLYQ MKEYGIYAAS IAIISLIVEF DIKRRQAETL SVK LMHRTA LTLCEKIRHL LVQKLQDMTY DDDDDNVNTE EVIMNFSTPK VQRFLMSLKV SFADKDPKDI CCLVFVERRY TCKC IYGLL LNYIQSTPEL RNVLTPQFMV GRNNISPDFE SVLERKWQKS AIQQFRDGNA NLMICSSVLE EGIDVQACNH VFILD PVKT FNMYVQSKGR ARTTEAKFVL FTADKEREKT IQQIYQYRKA HNDIAEYLKD RVLEKTEPEL YEIKGHFQDD IDPFTN ENG AVLLPNNALA ILHRYCQTIP TDAFGFVIPW FHVLQEDERD RIFGVSAKGK HVISINMPVN CMLRDTIYSD PMDNVKT AK ISAAFKACKV LYSLGELNER FVPKTLKERV ASIADVHFEH WNKYGDSVTA TVNKADKSKD RTYKTECPLE FYDALPRV G EICYAYEIFL EPQFESCEYT EHMYLNLQTP RNYAILLRNK LPRLAEMPLF SNQGKLHVRV ANAPLEVIIQ NSEQLELLH QFHGMVFRDI LKIWHPFFVL DRRSKENSYL VVPLILGAGE QKCFDWELMT NFRRLPQSHG SNVQQREQQP APRPEDFEGK IVTQWYANY DKPMLVTKVH RELTPLSYME KNQQDKTYYE FTMSKYGNRI GDVVHKDKFM IEVRDLTEQL TFYVHNRGKF N AKSKAKMK VILIPELCFN FNFPGDLWLK LIFLPSILNR MYFLLHAEAL RKRFNTYLNL HLLPFNGTDY MPRPLEIDYS LK RNVDPLG NVIPTEDIEE PKSLLEPMPT KSIEASVANL EITEFENPWQ KYMEPVDLSR NLLSTYPVEL DYYYHFSVGN VCE MNEMDF EDKEYWAKNQ FHMPTGNIYG NRTPAKTNAN VPALMPSKPT VRGKVKPLLI LQKTVSKEHI TPAEQGEFLA AITA SSAAD VFDMERLEIL GNSFLKLSAT LYLASKYSDW NEGTLTEVKS KLVSNRNLLF CLIDADIPKT LNTIQFTPRY TWLPP GISL PHNVLALWRE NPEFAKIIGP HNLRDLALGD EESLVKGNCS DINYNRFVEG CRANGQSFYA GADFSSEVNF CVGLVT IPN KVIADTLEAL LGVIVKNYGL QHAFKMLEYF KICRADIDKP LTQLLNLELG GKKMRANVNT TEIDGFLINH YYLEKNL GY TFKDRRYLLQ ALTHPSYPTN RITGSYQELE FIGNAILDFL ISAYIFENNT KMNPGALTDL RSALVNNTTL ACICVRHR L HFFILAENAK LSEIISKFVN FQESQGHRVT NYVRILLEEA DVQPTPLDLD DELDMTELPH ANKCISQEAE KGVPPKGEF NMSTNVDVPK ALGDVLEALI AAVYLDCRDL QRTWEVIFNL FEPELQEFTR KVPINHIRQL VEHKHAKPVF SSPIVEGETV MVSCQFTCM EKTIKVYGFG SNKDQAKLSA AKHALQQLSK CDA UniProtKB: Endoribonuclease Dcr-2 |
-Macromolecule #2: Loquacious, isoform D
| Macromolecule | Name: Loquacious, isoform D / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.502574 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDQENFHGSS LPQQLQNLHI QPQQASPNPV QTGFAPRRHY NNLVGLGNGN AVSGSPVKGA PLGQRHVKLK KEKISAQVAQ LSQPGQLQL SDVGDPALAG GSGLQGGVGL MGVILPSDEA LKFVSETDAN GLAMKTPVSI LQELLSRRGI TPGYELVQIE G AIHEPTFR ...String: MDQENFHGSS LPQQLQNLHI QPQQASPNPV QTGFAPRRHY NNLVGLGNGN AVSGSPVKGA PLGQRHVKLK KEKISAQVAQ LSQPGQLQL SDVGDPALAG GSGLQGGVGL MGVILPSDEA LKFVSETDAN GLAMKTPVSI LQELLSRRGI TPGYELVQIE G AIHEPTFR FRVSFKDKDT PFTAMGAGRS KKEAKHAAAR ALIDKLIGAQ LPESPSSSAG PSVTGLTVAG SGGDGNANAT GG GDASDKT VGNPIGWLQE MCMQRRWPPP SYETETEVGL PHERLFTIAC SILNYREMGK GKSKKIAKRL AAHRMWMRLQ ETP IDSGKI SDSICGELEG EVSIIQDIDR YEQVSKDFEF IKI UniProtKB: Protein Loquacious |
-Macromolecule #3: LD06392p
| Macromolecule | Name: LD06392p / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.115254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDNKSAVSAL QEFCARTQIN LPTYSFIPGE DGGYVCKVEL LEIEALGNGR SKRDAKHLAA SNILRKIQLL PGIHGLMKDS TVGDLDEEL TNLNRDMVKE LRDYCVRREM PLPCIEVVQQ SGTPSAPEFV ACCSVASIVR YGKSDKKKDA RQRAAIEMLA L ISSNSDNL ...String: MDNKSAVSAL QEFCARTQIN LPTYSFIPGE DGGYVCKVEL LEIEALGNGR SKRDAKHLAA SNILRKIQLL PGIHGLMKDS TVGDLDEEL TNLNRDMVKE LRDYCVRREM PLPCIEVVQQ SGTPSAPEFV ACCSVASIVR YGKSDKKKDA RQRAAIEMLA L ISSNSDNL RPDQMQVAST SKLKVVDMEE SMEELEALRR KKFTTYWELK EAGSVDHTGM RLCDRHNYFK NFYPTLKKEA IE AINSDEY ESSKDKAMDV MSSLKITPKI SEVESSSLVP LLSVELNCAF DVVLMAKETD IYDHIIDYFR TMLI UniProtKB: LD06392p |
-Macromolecule #4: RNA (52-MER)
| Macromolecule | Name: RNA (52-MER) / type: rna / ID: 4 / Details: dsRNA / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.646871 KDa |
| Sequence | String: GAGACUUGGG CAAUGUGACU GCUGAUCAGC AGUCACAUUG CCCAAGUCUC UU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Number classes used: 1 / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 3.72 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2) / Number images used: 141544 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8hf0: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)