[English] 日本語
 Yorodumi
Yorodumi- EMDB-34688: Local refinement of the SARS-CoV-2 Spike trimer in complex with R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEIN-IMMUNE SYSTEM COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Guo H / Gao Y / Lu Y / Yang H / Ji X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2023 Journal: Biochem Biophys Res Commun / Year: 2023Title: Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2. Authors: Xiaoyu Chu / Xinyu Ding / Yixuan Yang / Yuchi Lu / Tinghan Li / Yan Gao / Le Zheng / Hang Xiao / Tingting Yang / Hao Cheng / Haibin Huang / Yang Liu / Yang Lou / Chao Wu / Yuxin Chen / ...Authors: Xiaoyu Chu / Xinyu Ding / Yixuan Yang / Yuchi Lu / Tinghan Li / Yan Gao / Le Zheng / Hang Xiao / Tingting Yang / Hao Cheng / Haibin Huang / Yang Liu / Yang Lou / Chao Wu / Yuxin Chen / Haitao Yang / Xiaoyun Ji / Hangtian Guo /  Abstract: The COVID-19 pandemic, caused by SARS-CoV-2, has led to over 750 million infections and 6.8 million deaths worldwide since late 2019. Due to the continuous evolution of SARS-CoV-2, many significant ...The COVID-19 pandemic, caused by SARS-CoV-2, has led to over 750 million infections and 6.8 million deaths worldwide since late 2019. Due to the continuous evolution of SARS-CoV-2, many significant variants have emerged, creating ongoing challenges to the prevention and treatment of the pandemic. Therefore, the study of antibody responses against SARS-CoV-2 is essential for the development of vaccines and therapeutics. Here we perform single particle cryo-electron microscopy (cryo-EM) structure determination of a rabbit monoclonal antibody (RmAb) 9H1 in complex with the SARS-CoV-2 wild-type (WT) spike trimer. Our structural analysis shows that 9H1 interacts with the receptor-binding motif (RBM) region of the receptor-binding domain (RBD) on the spike protein and by directly competing with angiotensin-converting enzyme 2 (ACE2), it blocks the binding of the virus to the receptor and achieves neutralization. Our findings suggest that utilizing rabbit-derived mAbs provides valuable insights into the molecular interactions between neutralizing antibodies and spike proteins and may also facilitate the development of therapeutic antibodies and expand the antibody library. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34688.map.gz emd_34688.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34688-v30.xml emd-34688-v30.xml emd-34688.xml emd-34688.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34688.png emd_34688.png | 24.8 KB | ||
| Filedesc metadata |  emd-34688.cif.gz emd-34688.cif.gz | 6.4 KB | ||
| Others |  emd_34688_half_map_1.map.gz emd_34688_half_map_1.map.gz emd_34688_half_map_2.map.gz emd_34688_half_map_2.map.gz | 475.7 MB 475.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34688 http://ftp.pdbj.org/pub/emdb/structures/EMD-34688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34688 | HTTPS FTP |
-Validation report
| Summary document |  emd_34688_validation.pdf.gz emd_34688_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34688_full_validation.pdf.gz emd_34688_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_34688_validation.xml.gz emd_34688_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_34688_validation.cif.gz emd_34688_validation.cif.gz | 22.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34688 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34688 | HTTPS FTP |
-Related structure data
| Related structure data |  8hedMC  8hebC  8hecC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34688.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34688.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34688_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
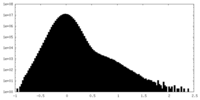
| Projections & Slices |
| ||||||||||||
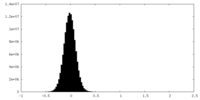
| Density Histograms |
-Half map: #1
| File | emd_34688_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
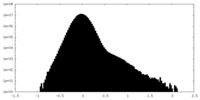
| Projections & Slices |
| ||||||||||||
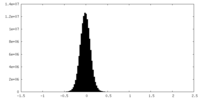
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 WT spike glycoprotein RBD complex with 9H1 Fabs
| Entire | Name: SARS-CoV-2 WT spike glycoprotein RBD complex with 9H1 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 WT spike glycoprotein RBD complex with 9H1 Fabs
| Supramolecule | Name: SARS-CoV-2 WT spike glycoprotein RBD complex with 9H1 Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 spike glycoprotein RBD
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein RBD / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: SARS-CoV-2 WT spike protein receptor-binding domain |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 9H1 Fab
| Supramolecule | Name: 9H1 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.446193 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLYRLFRKS NLKPFERDIS TEIYQAGSTP CNGVEGFNCY F PLQSYGFQ ...String: NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLYRLFRKS NLKPFERDIS TEIYQAGSTP CNGVEGFNCY F PLQSYGFQ PTNGVGYQPY RVVVLSFELL HAPATVCGPK KS UniProtKB: Spike glycoprotein |
-Macromolecule #2: rabbit antibody 9H1 light chain
| Macromolecule | Name: rabbit antibody 9H1 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.65484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVLTQTPSSV SAAVGGTVTI NCQASEDIYD NLVWYQQKPG QPPKLLIYDA STLAFGVSSR FRGSGSGTHF TLTMRDVQCD DAATYYCQG EFSCSSGDCT AFGGGTEVVV K |
-Macromolecule #3: rabbit antibody 9H1 heavy chain
| Macromolecule | Name: rabbit antibody 9H1 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.556056 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVEESGGRL VTPGTPLTLT CTVSGFSLSR YAMSWVRQAP GKGLEWIGII VDSGHTAYAS WAKGRFTISR TSTTVDLKMT SLTTEDTAT YFCARETGGG AFYVFEFWGP GTVVTVSS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X