[English] 日本語
 Yorodumi
Yorodumi- EMDB-34631: Focus refined map of C-lobe of human soluble guanylate cyclase in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focus refined map of C-lobe of human soluble guanylate cyclase in the NO+Rio state at 3.1 angstrom | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Chen L / Liu R | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nitric Oxide / Year: 2023 Journal: Nitric Oxide / Year: 2023Title: NO binds to the distal site of haem in the fully activated soluble guanylate cyclase. Authors: Rui Liu / Yunlu Kang / Lei Chen /  Abstract: Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO). The binding of NO to the haem of sGC induces a large conformational change in the enzyme and activates its cyclase ...Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO). The binding of NO to the haem of sGC induces a large conformational change in the enzyme and activates its cyclase activity. However, whether NO binds to the proximal site or the distal site of haem in the fully activated state remains under debate. Here, we present cryo-EM maps of sGC in the NO-activated state at high resolutions, allowing the observation of the density of NO. These cryo-EM maps show the binding of NO to the distal site of haem in the NO-activated state. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34631.map.gz emd_34631.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34631-v30.xml emd-34631-v30.xml emd-34631.xml emd-34631.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34631.png emd_34631.png | 35.1 KB | ||
| Masks |  emd_34631_msk_1.map emd_34631_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_34631_additional_1.map.gz emd_34631_additional_1.map.gz emd_34631_half_map_1.map.gz emd_34631_half_map_1.map.gz emd_34631_half_map_2.map.gz emd_34631_half_map_2.map.gz | 31.9 MB 59.4 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34631 http://ftp.pdbj.org/pub/emdb/structures/EMD-34631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34631 | HTTPS FTP |
-Validation report
| Summary document |  emd_34631_validation.pdf.gz emd_34631_validation.pdf.gz | 784.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34631_full_validation.pdf.gz emd_34631_full_validation.pdf.gz | 783.7 KB | Display | |
| Data in XML |  emd_34631_validation.xml.gz emd_34631_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_34631_validation.cif.gz emd_34631_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34631 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34631 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34631.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34631.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34631_msk_1.map emd_34631_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
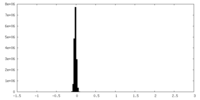
| Density Histograms |
-Additional map: #1
| File | emd_34631_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
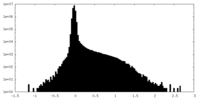
| Density Histograms |
-Half map: #1
| File | emd_34631_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
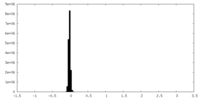
| Density Histograms |
-Half map: #2
| File | emd_34631_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
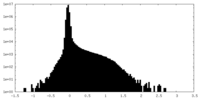
| Density Histograms |
- Sample components
Sample components
-Entire : human soluble guanylate cyclase
| Entire | Name: human soluble guanylate cyclase |
|---|---|
| Components |
|
-Supramolecule #1: human soluble guanylate cyclase
| Supramolecule | Name: human soluble guanylate cyclase / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 330636 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)