+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | NARROW LEAF 1-close from Japonica | |||||||||
 Map data Map data | Electron Microscopy Volume Map of Protein NARROW LEAF 1. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rice / panicle shape / photosynthetic efficiency / auxin transport / SURFACTANT PROTEIN | |||||||||
| Function / homology | stem vascular tissue pattern formation / internode patterning / regulation of leaf development / leaf vascular tissue pattern formation / Peptidase S1, PA clan / nucleoplasm / cytoplasm / Protein NARROW LEAF 1 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Zhang SJ / He YJ / Wang N / Zhang WJ / Liu CM | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: NARROW LEAF 1-close from Japonica Authors: Zhang SJ / He YJ / Wang N / Zhang WJ / Liu CM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34607.map.gz emd_34607.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34607-v30.xml emd-34607-v30.xml emd-34607.xml emd-34607.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34607.png emd_34607.png | 40.2 KB | ||
| Masks |  emd_34607_msk_1.map emd_34607_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34607.cif.gz emd-34607.cif.gz | 6.8 KB | ||
| Others |  emd_34607_half_map_1.map.gz emd_34607_half_map_1.map.gz emd_34607_half_map_2.map.gz emd_34607_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34607 http://ftp.pdbj.org/pub/emdb/structures/EMD-34607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34607 | HTTPS FTP |
-Related structure data
| Related structure data |  8hasMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34607.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34607.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron Microscopy Volume Map of Protein NARROW LEAF 1. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0738 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34607_msk_1.map emd_34607_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
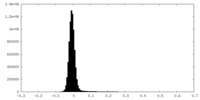
| Density Histograms |
-Half map: Electron Microscopy Half Map A of Protein NARROW LEAF 1.
| File | emd_34607_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron Microscopy Half Map A of Protein NARROW LEAF 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
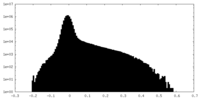
| Density Histograms |
-Half map: Electron Microscopy Half Map B of Protein NARROW LEAF 1.
| File | emd_34607_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron Microscopy Half Map B of Protein NARROW LEAF 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NARROW LEAF 1-close from Japonica
| Entire | Name: NARROW LEAF 1-close from Japonica |
|---|---|
| Components |
|
-Supramolecule #1: NARROW LEAF 1-close from Japonica
| Supramolecule | Name: NARROW LEAF 1-close from Japonica / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 379.51 kDa/nm |
-Macromolecule #1: Protein NARROW LEAF 1
| Macromolecule | Name: Protein NARROW LEAF 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.317902 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKPSDDKAQL SGLAQSEESS LDVDHQSFPC SPSIQPVASG CTHTENSAAY FLWPTSNLQH CAAEGRANYF GNLQKGLLPR HPGRLPKGQ QANSLLDLMT IRAFHSKILR RFSLGTAVGF RIRKGDLTDI PAILVFVARK VHKKWLNPAQ CLPAILEGPG G VWCDVDVV ...String: MKPSDDKAQL SGLAQSEESS LDVDHQSFPC SPSIQPVASG CTHTENSAAY FLWPTSNLQH CAAEGRANYF GNLQKGLLPR HPGRLPKGQ QANSLLDLMT IRAFHSKILR RFSLGTAVGF RIRKGDLTDI PAILVFVARK VHKKWLNPAQ CLPAILEGPG G VWCDVDVV EFSYYGAPAQ TPKEQMFSEL VDKLCGSDEC IGSGSQVASH ETFGTLGAIV KRRTGNKQVG FLTNHHVAVD LD YPNQKMF HPLPPNLGPG VYLGAVERAT SFITDDVWYG IYAGTNPETF VRADGAFIPF ADDFDISTVT TVVRGVGDIG DVK VIDLQC PLNSLIGRQV CKVGRSSGHT TGTVMAYALE YNDEKGICFF TDILVVGENR QTFDLEGDSG SLIILTSQDG EKPR PIGII WGGTANRGRL KLTSDHGPEN WTSGVDLGRL LDRLELDIII TNESLQDAVQ QQRFALVAAV TSAVGESSGV PVAIP EEKI EEIFEPLGIQ IQQLPRHDVA ASGTEGEEAS NTVVNVEEHQ FISNFVGMSP VRDDQDAPRS ITNLNNPSEE ELAMSL HLG DREPKRLRSD SGSSLDLEK UniProtKB: Protein NARROW LEAF 1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: The grid was coated with gold prior to use |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Vitrification carried out in Ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3770 / Average exposure time: 0.972 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 22500 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial local fitting was done using ChimeraX |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 153.3 |
| Output model |  PDB-8has: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)