+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Capsid structure of Ralstonia phage GP4 | |||||||||||||||
 Map data Map data | capsid | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ralstonia phage GP4 / Complex / VIRUS | |||||||||||||||
| Function / homology | Protein of unknown function DUF4043 / Phage capsid protein / Virion associated protein / Major capsid protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Liu HR / Chen WY | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: A Capsid Structure of GP4 with a Triangulation Number T = 9. Authors: Jing Zheng / Wenyuan Chen / Hao Xiao / Fan Yang / Xiaowu Li / Jingdong Song / Lingpeng Cheng / Hongrong Liu /  Abstract: GP4, a new phage, is a short-tailed phage. Few structures of phages have been resolved to near-atomic resolution until now. Here, we present a 3.7 Å resolution structure of the GP4 head by cryo- ...GP4, a new phage, is a short-tailed phage. Few structures of phages have been resolved to near-atomic resolution until now. Here, we present a 3.7 Å resolution structure of the GP4 head by cryo-electron microscopy (cryo-EM). The GP4 head contains 540 copies of major capsid protein (MCP) gp2 and 540 copies of cement protein (CP) gp1 arranged in an icosahedral shell with a triangulation number T = 9. The structures of gp2 and gp1 show a canonical HK97-like fold and an Ig-like fold, respectively. The trimeric CPs stick on the surface of the head along the quasi-threefold axis of the icosahedron generating a sandwiched three-layer electrostatic complementary potential, thereby enhancing the head stability. The assembly pattern of the GP4 head provides a platform for the further exploration of the interaction between and corresponding phages. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34539.map.gz emd_34539.map.gz | 858.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34539-v30.xml emd-34539-v30.xml emd-34539.xml emd-34539.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34539.png emd_34539.png | 144.7 KB | ||
| Others |  emd_34539_half_map_1.map.gz emd_34539_half_map_1.map.gz emd_34539_half_map_2.map.gz emd_34539_half_map_2.map.gz | 225.6 MB 225.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34539 http://ftp.pdbj.org/pub/emdb/structures/EMD-34539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34539 | HTTPS FTP |
-Validation report
| Summary document |  emd_34539_validation.pdf.gz emd_34539_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34539_full_validation.pdf.gz emd_34539_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_34539_validation.xml.gz emd_34539_validation.xml.gz | 22.4 KB | Display | |
| Data in CIF |  emd_34539_validation.cif.gz emd_34539_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34539 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34539 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34539 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34539 | HTTPS FTP |
-Related structure data
| Related structure data |  8h89MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34539.map.gz / Format: CCP4 / Size: 926.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34539.map.gz / Format: CCP4 / Size: 926.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | capsid | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 1
| File | emd_34539_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
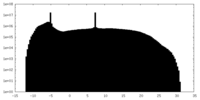
| Projections & Slices |
| ||||||||||||
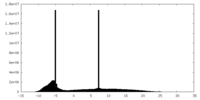
| Density Histograms |
-Half map: half map 2
| File | emd_34539_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
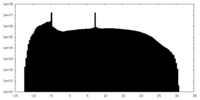
| Projections & Slices |
| ||||||||||||
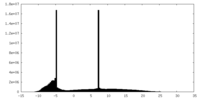
| Density Histograms |
- Sample components
Sample components
-Entire : Ralstonia phage GP4
| Entire | Name:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Ralstonia phage GP4
| Supramolecule | Name: Ralstonia phage GP4 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2282904 / Sci species name: Ralstonia phage GP4 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
| Molecular weight | Theoretical: 40.819551 KDa |
| Sequence | String: MSSTVIAFGD PKAQKKWSSE LAVDIRKKSY FESRFIGTSE NAVIQRKTEV ESDAGDRVSF DLSVRLRGQP TFGDDRVEGK EENLKFYTD EVIIDQVRHS VSAGGRMSRK RTAHDLRKTG RDRLGDYFYQ LTDELFFMYL SGARGINKDF ILPTSFTGYA K NPFNTPDA ...String: MSSTVIAFGD PKAQKKWSSE LAVDIRKKSY FESRFIGTSE NAVIQRKTEV ESDAGDRVSF DLSVRLRGQP TFGDDRVEGK EENLKFYTD EVIIDQVRHS VSAGGRMSRK RTAHDLRKTG RDRLGDYFYQ LTDELFFMYL SGARGINKDF ILPTSFTGYA K NPFNTPDA AHLLYGGVAT SKASLANTDT MSRVVIERAN VQATMMQAQD PETANMVPVS VEGEDRYVCV MSPFQEHSLR TS DAAGWLE IQKAAAAAEG RNNPIFKGGL GMIGNTVLHS HRNVVRFSDY GAGSDQPAAR ALFMGRQAAV VAYGTKGGLR YDW QEETKD YGNEPTVASG FIAGIKKTRF NDRDFGVISI DTYAKDPNPN NPA UniProtKB: Major capsid protein |
-Macromolecule #2: Virion associated protein
| Macromolecule | Name: Virion associated protein / type: protein_or_peptide / ID: 2 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ralstonia phage GP4 (virus) Ralstonia phage GP4 (virus) |
| Molecular weight | Theoretical: 15.819934 KDa |
| Sequence | String: MALIQSDFAQ GIRMTPVPDC AGDVTACRFD ITLKNAPAAG DIIELGVLPG NAVPVEAILD VDDLDTGGAP TITLDVGIMS GPVGKNDPA RTCGNELFAA STVGQAGGVV RATASSAFRI QKAEDHRSVG VKVAAGPATG AAGKTIALIL FYVQGTSQ UniProtKB: Virion associated protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 0.1 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 40792 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)