+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
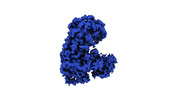
| Title | The core region map of avian influenza A polymerase. | ||||||||||||
 Map data Map data | The core region map of avian influenza A polymerase. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Polymerase / VIRAL PROTEIN | ||||||||||||
| Biological species |   Influenza A virus Influenza A virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Li H / Wu Y / Liang H / Liu Y | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles. Authors: Huanhuan Li / Yixi Wu / Minke Li / Lu Guo / Yaqi Gao / Quan Wang / Jihua Zhang / Zhaohua Lai / Xing Zhang / Lixin Zhu / Ping Lan / Zihe Rao / Yingfang Liu / Huanhuan Liang /  Abstract: Influenza polymerase (FluPol) transcribes viral mRNA at the beginning of the viral life cycle and initiates genome replication after viral protein synthesis. However, it remains poorly understood how ...Influenza polymerase (FluPol) transcribes viral mRNA at the beginning of the viral life cycle and initiates genome replication after viral protein synthesis. However, it remains poorly understood how FluPol switches between its transcription and replication states, especially given that the structural bases of these two functions are fundamentally different. Here we propose a mechanism by which FluPol achieves functional switching between these two states through a previously unstudied conformation, termed an 'intermediate state'. Using cryo-electron microscopy, we obtained a structure of the intermediate state of H5N1 FluPol at 3.7 Å, which is characterized by a blocked cap-binding domain and a contracted core region. Structural analysis results suggest that the intermediate state may allow FluPol to transition smoothly into either the transcription or replication state. Furthermore, we show that the formation of the intermediate state is required for both the transcription and replication activities of FluPol, leading us to conclude that the transcription and replication cycles of FluPol are regulated via this intermediate state. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34496.map.gz emd_34496.map.gz | 4.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34496-v30.xml emd-34496-v30.xml emd-34496.xml emd-34496.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34496.png emd_34496.png | 33.6 KB | ||
| Others |  emd_34496_half_map_1.map.gz emd_34496_half_map_1.map.gz emd_34496_half_map_2.map.gz emd_34496_half_map_2.map.gz | 13.3 MB 13.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34496 http://ftp.pdbj.org/pub/emdb/structures/EMD-34496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34496 | HTTPS FTP |
-Validation report
| Summary document |  emd_34496_validation.pdf.gz emd_34496_validation.pdf.gz | 510.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34496_full_validation.pdf.gz emd_34496_full_validation.pdf.gz | 509.7 KB | Display | |
| Data in XML |  emd_34496_validation.xml.gz emd_34496_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_34496_validation.cif.gz emd_34496_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34496 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34496 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34496.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34496.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The core region map of avian influenza A polymerase. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: The half1 map of core region of avian influenza A polymerase.
| File | emd_34496_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half1 map of core region of avian influenza A polymerase. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The half2 map of core region of avian influenza A polymerase.
| File | emd_34496_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half2 map of core region of avian influenza A polymerase. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza A virus RdRp complex
| Entire | Name: Influenza A virus RdRp complex |
|---|---|
| Components |
|
-Supramolecule #1: Influenza A virus RdRp complex
| Supramolecule | Name: Influenza A virus RdRp complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 8.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 779360 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)