+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a riboendonuclease | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | riboendonuclease / RNA | |||||||||
| Function / homology | : / endonuclease activity / defense response to virus / RNA binding / CRISPR-associated endoribonuclease Cas13a Function and homology information Function and homology information | |||||||||
| Biological species |  Thermoclostridium caenicola (bacteria) Thermoclostridium caenicola (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Li Z / Wang F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola. Authors: Feng Wang / Chendi Zhang / Haijiang Xu / Wanting Zeng / Lixin Ma / Zhuang Li /  Abstract: The RNA-targeting type VI CRISPR-Cas effector complexes are widely used in biotechnology applications such as gene knockdown, RNA editing, and molecular diagnostics. Compared with Cas13a from ...The RNA-targeting type VI CRISPR-Cas effector complexes are widely used in biotechnology applications such as gene knockdown, RNA editing, and molecular diagnostics. Compared with Cas13a from mesophilic organisms, a newly discovered Cas13a from thermophilic bacteria Thermoclostridium caenicola (TccCas13a) shows low sequence similarity, high thermostability, and lacks pre-crRNA processing activity. The thermostability of TccCas13a has been harnessed to make a sensitive and robust tool for nucleic acid detection. Here we present the structures of TccCas13a-crRNA binary complex at 2.8 Å, and TccCas13a at 3.5 Å. Although TccCas13a shares a similarly bilobed architecture with other mesophilic organism-derived Cas13a proteins, TccCas13a displayed distinct structure features. Specifically, it holds a long crRNA 5'-flank, forming extensive polar contacts with Helical-1 and HEPN2 domains. The detailed analysis of the interaction between crRNA 5'-flank and TccCas13a suggested lack of suitable nucleophile to attack the 2'-OH of crRNA 5'-flank may explain why TccCas13a fails to cleave pre-crRNA. The stem-loop segment of crRNA spacer toggles between double-stranded and single-stranded conformational states, suggesting a potential safeguard mechanism for target recognition. Superimposition of the structures of TccCas13a and TccCas13a-crRNA revealed several conformational changes required for crRNA loading, including dramatic movement of Helical-2 domain. Collectively, these structural insights expand our understanding into type VI CRISPR-Cas effectors, and would facilitate the development of TccCas13a-based applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34484.map.gz emd_34484.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34484-v30.xml emd-34484-v30.xml emd-34484.xml emd-34484.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
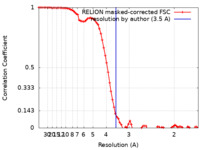
| FSC (resolution estimation) |  emd_34484_fsc.xml emd_34484_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34484.png emd_34484.png | 92.4 KB | ||
| Filedesc metadata |  emd-34484.cif.gz emd-34484.cif.gz | 6.4 KB | ||
| Others |  emd_34484_half_map_1.map.gz emd_34484_half_map_1.map.gz emd_34484_half_map_2.map.gz emd_34484_half_map_2.map.gz | 31.4 MB 31.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34484 http://ftp.pdbj.org/pub/emdb/structures/EMD-34484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34484 | HTTPS FTP |
-Validation report
| Summary document |  emd_34484_validation.pdf.gz emd_34484_validation.pdf.gz | 695.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34484_full_validation.pdf.gz emd_34484_full_validation.pdf.gz | 694.8 KB | Display | |
| Data in XML |  emd_34484_validation.xml.gz emd_34484_validation.xml.gz | 13.6 KB | Display | |
| Data in CIF |  emd_34484_validation.cif.gz emd_34484_validation.cif.gz | 19.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34484 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34484 | HTTPS FTP |
-Related structure data
| Related structure data |  8h4uMC  8ewgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34484.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34484.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
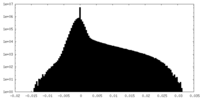
| Projections & Slices |
| ||||||||||||
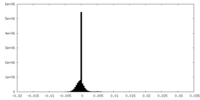
| Density Histograms |
-Half map: #1
| File | emd_34484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
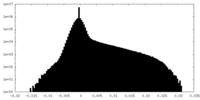
| Projections & Slices |
| ||||||||||||
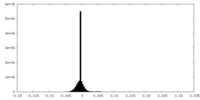
| Density Histograms |
- Sample components
Sample components
-Entire : Apo-form riboendonuclease
| Entire | Name: Apo-form riboendonuclease |
|---|---|
| Components |
|
-Supramolecule #1: Apo-form riboendonuclease
| Supramolecule | Name: Apo-form riboendonuclease / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thermoclostridium caenicola (bacteria) Thermoclostridium caenicola (bacteria) |
-Macromolecule #1: CRISPR-associated endonuclease Cas9
| Macromolecule | Name: CRISPR-associated endonuclease Cas9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoclostridium caenicola (bacteria) Thermoclostridium caenicola (bacteria) |
| Molecular weight | Theoretical: 143.582641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKITKRKWGE HHPPLYFYRD EDSGRLLAQN DRKQDYTDTL FNDIAQDTFE RSLRNRLLKT PEKGDKRFYS NEIVKLVEKL CQGADVAEI MKSMERNEKL RPKNEKEIKN LKKQLDGTLS EYGKRYTAPE GAMTLNDALF YLVEGNPLKQ AMAKAELGKI R EALIKEKE ...String: MKITKRKWGE HHPPLYFYRD EDSGRLLAQN DRKQDYTDTL FNDIAQDTFE RSLRNRLLKT PEKGDKRFYS NEIVKLVEKL CQGADVAEI MKSMERNEKL RPKNEKEIKN LKKQLDGTLS EYGKRYTAPE GAMTLNDALF YLVEGNPLKQ AMAKAELGKI R EALIKEKE NRINRVRYSI KNNKIPLRIQ EDGGITPNND RAAWLLGLMK PADPAKGITD CYPLLGELEE VFDFDKLSKT LH EKISRCQ GRPRSIAMAV DEALKQYLRE LWEKSPSRQQ DLKYYFQAVQ EYFKDNFPIR TKRMGARLRQ ELLKDKTSLS RLL EPKHMA NAVRRRLINQ STQMHILYGK LYAYCCGEDG RLLVNSETLQ RIQVHEAVKK QAMTAVLWSI SRLRYFYQFE DGDI LSNKN PIKDFRDKFL RDTNKYTHED VEACKEKLQD FFPLKELQEK IKEDAKGLQE TDNKQADTTD FKAIGHIVRD DRKLC NQLL AECVSCIGEL RHHIFHYKNV TLIQALKRIA DKVKPEDLSV LRAIYLLDRR NLKKAFAKRI SSMNLPLYYR EDLLSR IFK KEGTAFFLYS AKIQMTPSFQ RVYERGKNLR REFECERMKA EASNGQNGQD GDRLKWFRQL AAGDSADTHF NWAVEAY AE SAADVENNVE FDTDVDAQRA LRNLLLLIYR HHFLPEVQKD ETLVTGKIHK VLERNRQLSE GQGPNQGKAH GYSVIEEL Y HEGMPLSDLM KQLQRRISET ERESRELAQE KTDYAQRFIL DIFAEAFNDF LEAHYGEEYL EIMSPRKDAE AAKKWVKES KTVDLKTSID EKEPEGHLLV LYPVLRLLDE RELGELQQQM IRYRTSLASW QGESNFSEEI RIAGQIEELT ELVKLTEPEP QFAEEVWGK RAKEAFEDFI EGNMKNYEAF YLQSDNNTPV YRRNMSRLLR SGLMGVYQKV LASHKQALKR DYLLWSEKHW N VKDENGAD ISSAEQAQCL LQRLHRKYAE SPSRFTEEDC KLYEKVLRRL EDYNQAVKNL SFSSLYEICV LNLEILSRWV GF VQDWERD MYFLLLAWVR QGKLDGIKEE DVRDIFSEGN IIRNLVDTLK GENMNAFESV YFPENKGSKY LGVRNDVAHL DLM RKNGWR LEAGKTCSVM EDYINRLRFL LSYDQKRMNA VTKTLQQIFD RHKVKIRFTV EKGGMLKIED VTADKIVHLK GSRL SGIEI PSHGERFIDT LKALMVYPRG UniProtKB: CRISPR-associated endoribonuclease Cas13a |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)