+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GW9508-bound FFAR4 in complex with Gq | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR-G-protein complex / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / hormone secretion / arrestin family protein binding / negative regulation of cytokine production / negative regulation of interleukin-1 beta production ...negative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / hormone secretion / arrestin family protein binding / negative regulation of cytokine production / negative regulation of interleukin-1 beta production / ciliary membrane / white fat cell differentiation / endocytic vesicle / positive regulation of osteoblast differentiation / brown fat cell differentiation / positive regulation of brown fat cell differentiation / fatty acid binding / G protein-coupled receptor activity / negative regulation of inflammatory response / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / positive regulation of ERK1 and ERK2 cascade / endosome membrane / ciliary basal body / cilium / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / intracellular membrane-bounded organelle / GTPase activity / synapse / negative regulation of apoptotic process / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
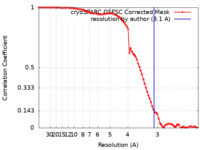
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | He Y / Yin H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity. Authors: Han Yin / Asuka Inoue / Zhengxiong Ma / Xinyan Zhu / Ruixue Xia / Zhenmei Xu / Na Wang / Yaning Duan / Anqi Zhang / Changyou Guo / Yuanzheng He /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34479.map.gz emd_34479.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34479-v30.xml emd-34479-v30.xml emd-34479.xml emd-34479.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34479_fsc.xml emd_34479_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_34479.png emd_34479.png | 78.7 KB | ||
| Filedesc metadata |  emd-34479.cif.gz emd-34479.cif.gz | 6.4 KB | ||
| Others |  emd_34479_half_map_1.map.gz emd_34479_half_map_1.map.gz emd_34479_half_map_2.map.gz emd_34479_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34479 http://ftp.pdbj.org/pub/emdb/structures/EMD-34479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34479 | HTTPS FTP |
-Validation report
| Summary document |  emd_34479_validation.pdf.gz emd_34479_validation.pdf.gz | 842.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34479_full_validation.pdf.gz emd_34479_full_validation.pdf.gz | 841.8 KB | Display | |
| Data in XML |  emd_34479_validation.xml.gz emd_34479_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_34479_validation.cif.gz emd_34479_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34479 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34479 | HTTPS FTP |
-Related structure data
| Related structure data |  8h4kMC  8h4iC  8h4lC  8iysC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34479.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34479.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
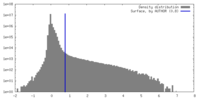
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34479_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
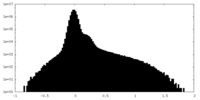
| Projections & Slices |
| ||||||||||||
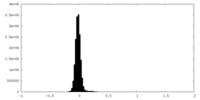
| Density Histograms |
-Half map: #1
| File | emd_34479_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
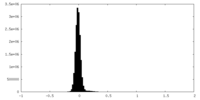
| Density Histograms |
- Sample components
Sample components
-Entire : GPCR/G-protein complex
| Entire | Name: GPCR/G-protein complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: engineered mini Galpha-Q subunit
| Macromolecule | Name: engineered mini Galpha-Q subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.855578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY ...String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY IPTQQDVLRT RVKTSGIFET KFQVDKVNFH MFDVGAQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL ND FKSIWNN RWLRTISVIL FLNKQDLLAE KVLAGKSKIE DYFPEFARYT TPEDATPEPG EDPRVTRAKY FIRKEFVDIS TAS GDGRHI CYPHFTCSVD TENARRIFND CKDIILQMNL REYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.381584 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGR FTISRDNAKN TLYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SAAALEHHHH H H |
-Macromolecule #5: Free fatty acid receptor 4
| Macromolecule | Name: Free fatty acid receptor 4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.533211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSPECARAAG DAPLRSLEQA NRTRFPFFSD VKGDHRLVLA AVETTVLVLI FAVSLLGNVC ALVLVARRRR RGATACLVLN LFCADLLFI SAIPLVLAVR WTEAWLLGPV ACHLLFYVMT LSGSVTILTL AAVSLERMVC IVHLQRGVRG PGRRARAVLL A LIWGYSAV ...String: MSPECARAAG DAPLRSLEQA NRTRFPFFSD VKGDHRLVLA AVETTVLVLI FAVSLLGNVC ALVLVARRRR RGATACLVLN LFCADLLFI SAIPLVLAVR WTEAWLLGPV ACHLLFYVMT LSGSVTILTL AAVSLERMVC IVHLQRGVRG PGRRARAVLL A LIWGYSAV AALPLCVFFR VVPQRLPGAD QEISICTLIW PTIPGEISWD VSFVTLNFLV PGLVIVISYS KILQITKASR KR LTVSLAY SESHQIRVSQ QDFRLFRTLF LLMVSFFIMW SPIIITILLI LIQNFKQDLV IWPSLFFWVV AFTFANSALN PIL YNMTLC RNEWKKIFCC FWFPEKGAIL TDTSVKRNDL SIISG UniProtKB: Free fatty acid receptor 4 |
-Macromolecule #6: 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid
| Macromolecule | Name: 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: WPT |
|---|---|
| Molecular weight | Theoretical: 347.407 Da |
| Chemical component information | 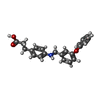 ChemComp-WPT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)